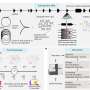
Want to better understand the accuracy of your human genome sequencing?

As new high-throughput "Next Generation" DNA sequencing methods are moving into clinical applications, understanding accuracy of variants is critical. Numerous recent studies have demonstrated that different sequencing and bioinformatics analysis methods can result in very different variant calls, often at 100,000's of positions across the human genome.
With help from the Genome in a Bottle Consortium (www.genomeinabottle.org) and the FDA, NIST is developing well-characterized whole human genomes as Reference Materials, as well as the methods to use these Reference Materials to understand performance of sequencing and bioinformatics methods, including false positive and false negative rates.
We recently released our first set of highly confident small variant (SNP and indel) genotypes for our pilot candidate Reference Material, based on the NA12878 DNA from Coriell (ccr.coriell.org). This characterization includes both highly confident variant calls, where our Reference Material is different from the reference genome assembly, and highly confident homozygous reference calls, where our Reference Material is the same as the reference genome assembly (see more details about using these in the blog post genomeinabottle.org/blog-entry … s-available-ftp-site .)
Research and clinical laboratories interested in understanding and improving performance of the sequencing and/or bioinformatics have started using our highly confident genotypes by comparing their variant calls to ours in our highly confident regions. They can look at genomic locations where they disagree to see how to improve their methods. Interested laboratories can either sequence NA12878 DNA from Coriell themselves or download data others have generated at our Genome in a Bottle ftp site at NCBI (see genomeinabottle.org/blog-entry … p-site-now-live-ncbi ).
In addition, we have collaborated with the developers of the GCAT website (www.bioplanet.com/gcat) to allow anyone to compare variant calls from different bioinformatics to our highly confident genotype in an interactive environment.
We also have ~8300 vials of DNA from a homogenized large batch of NA12878 cells, which we are starting to send to laboratories interested in helping us to characterize this candidate NIST Reference Material. After characterizing the stability and homogeneity, we expect to distribute this DNA as a NIST Reference Material. We also plan to develop well-characterized whole genome Reference Materials from an additional set of ~8 father-mother-child trios from diverse ancestry groups from the Personal Genome Project.