Discovery to help predict who will benefit from lung cancer treatment
Cancer Research UK scientists have discovered the structure of an abnormal protein which causes an aggressive type of lung cancer, according to new research* published in the Proceedings of the National Academy of Science on Monday.
Unveiling the structure of this protein – formed by a genetic fault – could enable doctors to predict who will benefit from a specific lung cancer treatment, while saving other patients from receiving it unnecessarily.
Researchers were looking at a form of the disease – known as ALK lung cancers – which account for around four per cent of cases.
These lung cancers have a fault where two different genes become locked together. This gene fusion forms a souped-up version of a protein which then becomes an engine, driving the cancer to grow very fast and spread rapidly.
But, importantly these cancers rely on this engine to survive so blocking the protein could kill the lung cancer.
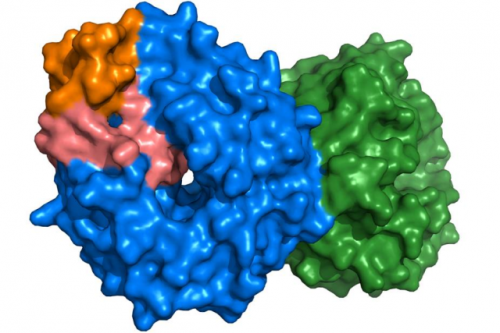
Using x-ray crystallography, the researchers were able to develop a clear picture of the shape of one half of the souped-up protein. The shape of the other half was already known. This revealed several different shapes depending on where the genes had fused.
Crucially, some of the shapes are unstable and need help from another protein to work. This assistant protein can be blocked by drugs known as Hsp90 inhibitors. By stopping this helper protein, the unstable, souped-up proteins can no longer work and the cancer cells die.
But for around one third of patients with ALK lung cancer, the structure of the protein uncovered by the researchers is much more stable and is resistant to the Hsp90 inhibitors.
Following on from their discovery, the researchers grew cells in the lab to test their theory. As predicted, the cells with the unstable protein were killed by the drug, and the cells with the stable form of the protein continued to grow.
The researchers are currently collecting data and samples from a clinical trial to find out if their lab findings hold true and can be used to predict which lung cancer patients will respond to the drug.
Dr Richard Bayliss, co-author based at the University of Leicester and the Cancer Research UK Leicester Centre, said: "We routinely use protein structures during the process of drug design, but this is the first time they have helped us to predict patient response to drugs in clinical development. The other unique aspect of this project has been the bringing together of teams who hadn't previously worked together. We had structural biologists working alongside clinical researchers who help treat patients. Our results would not have been possible without the clinical oncology expertise of Professor Dean Fennell and his team. We're now looking for groups of patients who might benefit from Hsp90 inhibitors because they harbour other 'souped-up' proteins with unstable shapes."
Dr Emma Smith, Cancer Research UK's senior science information officer, said: "This study is a positive step forward in making sure lung cancer patients get the most effective treatment based on the genetic mistakes that underpin their disease. We now need to build on this research and gather further clinical data to confirm these findings. It may lead to doctors developing a simple genetic test to spot patients who will benefit from a drug targeted against their disease, and spare patients unlikely to benefit unnecessary side effects.
"Lung cancer has been a difficult disease to overcome. Unravelling the mystery of its complex biology, and developing better and kinder treatments is taking time but this research provides yet more hope that we're moving in the right direction."
More information: Mark W. Richards, Edward W. P. Law, La'Verne P. Rennalls, Sara Busacca, Laura O'Regan, Andrew M. Fry, Dean A. Fennell, and Richard Bayliss. "Crystal structure of EML1 reveals the basis for Hsp90 dependence of oncogenic EML4-ALK by disruption of an atypical β-propeller domain." PNAS 2014 ; published ahead of print March 24, 2014, DOI: 10.1073/pnas.1322892111