Humans may have benefited from mobile genetic element insertions
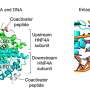
Mobile pieces of DNA can jump around in the human genome and exert powerful regulatory effects on neighboring genes at new insertion sites. The most common mobile genetic elements are known as LINE-1 retrotransposons and comprise almost 20 per cent of all our DNA. Yet most genome-wide studies have underestimated the full impact of these important genetic regulators.
To correct this, a team led by William Burkholder at the A*STAR Institute of Molecular and Cell Biology in Singapore took a fresh look at the effects of LINE-1 retrotransposons on gene function and disease susceptibility. When they did this, the researchers found evidence that a subset of LINE-1 elements seems to have exerted a beneficial effect on recent human evolution.
Alexandre Kuhn from the Burkholder laboratory and other colleagues performed a comprehensive survey of LINE-1 retrotransposons in the genomes of 20 individuals of Asian descent. The research team, which included scientists from Singapore and the United States, found that, on average, an individual has more than 800 LINE-1 retrotransposons. Despite their high prevalence, these important genetic elements have not been adequately accounted for in standard arrays designed for genome-wide analyses of human disease or natural variation.
The researchers showed that this oversight, rather than originating from a technical error, was due to a limitation of the most commonly used methodologies. Specifically, they found that the DNA letter differences, known as single-nucleotide polymorphisms (SNPs), measured on the arrays are not closely linked to nearby LINE-1 elements, and hence the standard arrays failed to 'tag' them.
In order to tag LINE-1 retrotransposons to neighboring SNPs, Burkholder's team integrated LINE-1 information with whole-genome sequencing data collected by the 1000 Genomes Project—a massive international effort to create a detailed map of human genetic variation. They found that most LINE-1 elements could indeed be tightly associated with SNPs. According to Kuhn, this means "that genome-wide association studies of their phenotypic effects could be conducted based on SNP genotyping in large human populations."
To prove the point, the researchers ran the new analysis and then studied the genetic sequences surrounding the mobile genetic elements. In this way, they found evidence for recent positive selection, meaning that some of the LINE-1 retrotransposons conferred an evolutionary advantage when they jumped into new spots in the genome. This could be seen in the unusually long blocks of DNA surrounding several LINE-1 retrotransposons—an indication that the genetic elements and their flanking sequences had swept through the population.
More information: Kuhn, A., Ong, Y. M., Cheng, C.-Y., Wong, T. Y., Quake, S. R. & Burkholder, W. F. Linkage disequilibrium and signatures of positive selection around LINE-1 retrotransposons in the human genome. Proceedings of the National Academy of Sciences USA 111, 8131–8136 (2014). dx.doi.org/10.1073/pnas.1401532111