Researchers bring order to big data of human biology
A multi-year study led by researchers from the Simons Center for Data Analysis (SCDA) and major universities and medical schools has broken substantial new ground, establishing how genes work together within 144 different human tissues and cell types in carrying out those tissues' functions.
The paper, to be published online by Nature Genetics on April 27, also demonstrates how computer science and statistical methods may combine to aggregate and analyze very large—and stunningly diverse—genomic 'big-data' collections.
Led by Olga Troyanskaya, deputy director for genomics at SCDA, the team collected and integrated data from about 38,000 genome-wide experiments (from an estimated 14,000 publications). These datasets necessarily contain not only information about cells' RNA/protein functions, but also information from individuals diagnosed with a variety of illnesses.
Using integrative computational analysis, the researchers first isolated the functional genetic interconnections contained in these rich datasets for various tissue types. Then, combining that tissue-specific functional signal with the relevant disease's DNA-based genome-wide association studies (GWAS), the researchers were able to identify statistical associations between genes and diseases that would otherwise be undetectable.
The resulting technique, which they called a 'network-guided association study,' or NetWAS, thus integrates quantitative genetics with functional genomics to increase the power of GWAS and identify genes underlying complex human diseases. And because the technique is completely data-driven, NetWAS avoids bias toward better-studied genes and pathways, permitting discovery of novel associations.
SCDA director Leslie Greengard says, "Olga and her collaborators have demonstrated that extraordinary results can be achieved by merging deep biological insight with state-of-the-art computational methods, and applying them to large-scale, noisy and heterogeneous datasets."
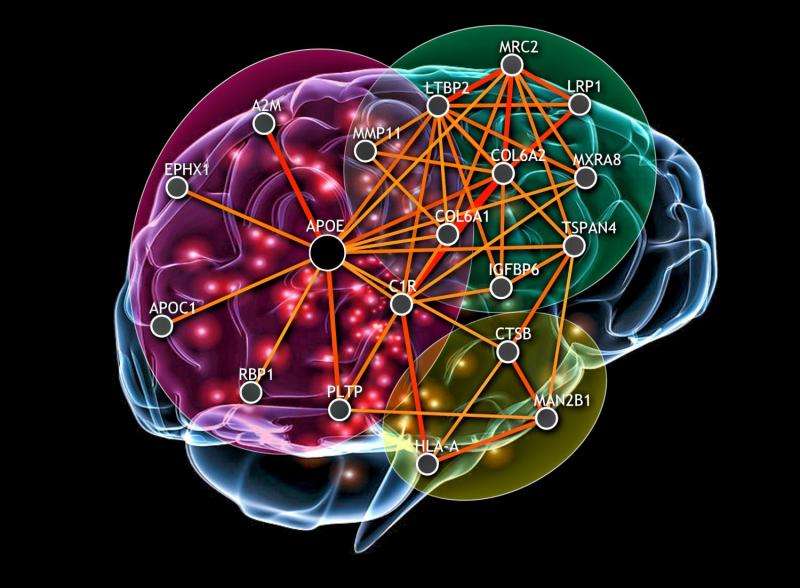
The result of their efforts was 144 functional gene interaction networks for organs as diverse as the kidney, the liver and the whole brain. The paper goes on to describe functional gene disruptions for diseases such as hypertension, diabetes and obesity.
Importantly, while such functional gene interaction networks had already been established in animal models, this feat had not yet been accomplished—and could not have been accomplished without 'big data'— in human tissue. Many human cell types important to disease cannot be studied by traditional direct experimentation, so the ability to instead work with these rich datasets was a critical workaround.
"A key challenge in human biology is that genetic circuits in human tissues and cell types are very difficult to study experimentally," says Troyanskaya, who also is a professor in the computer science department and the Lewis-Sigler Institute for Integrative Genomics at Princeton University. "For example, the podocyte cells in the kidneys that perform the kidney's filtering function cannot be isolated for study in the lab, nor can the function of genes be identified by genome-scale experiments. Yet we need to understand how proteins interact in these cells if we want to understand and treat chronic kidney disease. Our approach mined these big data collections to build a map of how genetic circuits function in the podocyte cells, and in many other disease-relevant tissues and cell types."
These findings have important implications for our understanding of normal gene function, but also for drug use and development: Causal or target genes may be better identified for treatment, and previously unexpected drug interactions and disruptions may be anticipated. "Biomedical researchers can use these networks and the pathways that they uncover to understand drug action and side effects in the context of specific disease-relevant tissues, and to repurpose drugs," Troyanskaya says. "These networks can also be useful for understanding how various therapies work and to help with developing new therapies."
The researchers have also created an online resource so that other scientists may use NetWAS and access the tissue-specific networks. The team created an interactive server, the Genome-scale Integrated Analysis of Networks in Tissues, or GIANT. GIANT allows users to explore the networks, compare how genetic circuits vary across tissues, and analyze data from genetic studies to find genes that cause disease.
Aaron K. Wong, a data scientist at SCDA and formerly a graduate student in the computer science department at Princeton, led the way in creating GIANT. "Our goal was to develop a resource that was accessible to biomedical researchers," he says. "For example, with GIANT, researchers studying Parkinson's disease can search the substantia nigra network, which represents the brain region affected by Parkinson's, to identify new genes and pathways involved in the disease." Wong is one of three co-first authors of the paper.
The paper's other two co-first authors are Arjun Krishnan, a postdoctoral fellow at the Lewis-Sigler Institute; and Casey S. Greene, assistant professor of genetics at Dartmouth College, who was a postdoctoral fellow with the Troyanskaya group from 2009 to 2012. Other key collaborators on this study were Emanuela Ricciotti, Garret A. FitzGerald and Tilo Grosser of the pharmacology department and the Institute for Translational Medicine and Therapeutics at the Perelman School of Medicine, University of Pennsylvania; Daniel I. Chasman of Brigham and Women's Hospital and Harvard Medical School in Boston; and Kara Dolinski at the Lewis-Sigler Institute at Princeton University.
"This is an exciting time in biomedical research, and I believe we are still at the early stages of developing new ways to think about biological networks and their control," Greengard says.
More information: Understanding multicellular function and disease with human tissue-specific networks, Nature Genetics, http://dx.doi.org/10.1038/ng.3259