Team develops method to better identify genes involved in diseases
Scientists at A*STAR's Genome Institute of Singapore (GIS) have developed a new technique that simplifies the task of identifying the precise DNA mutations that cause disease, which lays the groundwork for the development of new drugs and new ways of diagnosing diseases.
It is generally extremely difficult to design new drugs for diseases such as diabetes or rheumatoid arthritis (for example), because we do not know which molecules in the body the drugs should target. This is where disease-causing DNA mutations come in. If we can pinpoint the genetic mutations that cause a particular disease, we can associate those mutations with specific proteins, and then design drugs to block or activate those proteins. We can also measure the activity of proteins identified in this manner to diagnose diseases more accurately, which is a major goal of precision medicine.
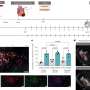
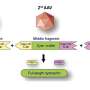
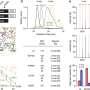
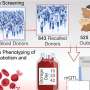
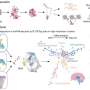
A team of scientists, led by Dr Shyam Prabhakar, Associate Director for Integrated Genomics at GIS, developed a new genetic analysis technique called the Genotype-independent Signal Correlation and Imbalance (G-SCI) test that senses specific chemical changes within the genome and connects them to nearby genetic mutations. They then showed that mutations associated with the chemical changes were also likely to cause disease. The G-SCI test was validated in a study of 57 individuals and is reported in the scientific journal, Nature Methods.
When combined with the chemical profiling strategy used in this study, the G-SCI test is 10 times more sensitive than existing methods at identifying detrimental gene mutations. The study's co-lead, Dr Ricardo del Rosario from GIS, said, "The G-SCI test is transformative – instead of examining gene expression correlations in 500 individuals, we can get away with histone acetylation analysis of a mere 50 to 60. This reduces the number of test subjects needed to conclude each study and gives us the ability to look into multiple diseases."
Dr Jeremie Poschmann from GIS, the other co-lead of the study, highlighted another benefit of the new approach: "Instead of using genome sequencing, we can use the histone acetylation sequencing data from our method to detect DNA mutations. This saves us a huge amount of time, effort and resources."
Prof Bing Ren of the Ludwig Institute of Cancer Research and the Department of Cellular and Molecular Medicine, University of California, San Diego (UCSD) said, "This is an exciting study that sets a new benchmark for genetic analysis of gene regulation. The method greatly enhances our ability to interpret the human genome and will benefit research into the genetic basis of diseases."
The scientists validated the efficiency of the new test and were able to identify links to certain genetic diseases. "We have found a strong association between mutations that perturbed the genome's chemical state and those that caused autoimmune diseases. That's when we knew we had hit the bulls-eye with the G-SCI test," said Dr Prabhakar.
"This work provides important new tools for linking genetic variation to variation in chromatin function, and provides compelling evidence for the central role of this type of genetic variation in human disease," commented Prof Jonathan Pritchard from Stanford University, who is also an Investigator at the Howard Hughes Medical Institute.
"I am thrilled by this new method with superior precision for identifying disease-causing mutations. The G-SCI method innovatively utilises epigenetic information to select regions of the human genome for disease association analysis. As we see bigger and more complex datasets, the community will face the forthcoming challenges of analysing big data. This method has expanded our arsenal of computational analytics capabilities at the Genome Institute of Singapore," said Prof Huck-Hui Ng, Executive Director of GIS.
More information: "Sensitive mapping of chromatin‐altering polymorphisms reveals molecular drivers of autoimmune disease" Nature Methods