October 2, 2015 report
Researchers identify genes associated with specific metabolic pathways

(Medical Xpress)—An international team of researchers working at the Washington University School of Medicine in Missouri has identified several genes associated with specific metabolic pathways in a mammal biome, including some that respond to a dramatic change in diet. In their paper published in the journal Science, the team describes a genetic study they conducted on four strains of gut bacteria and a secondary experiment involving genes associated with a change in diet.
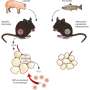
Scientists have known for a long time that the mammalian gut is more complicated than it seems—more recently, they have found that the gut biome is chock full of different species of bacteria, most of which help with digestion. One area of study in particular involves how the biome is able to adapt to a change in diet. In this new effort, the researchers sought to learn more about the process by studying a much simpler system, a mouse gut that had been sterilized and then implanted with just four human Bacteroides strains.
The idea was to see how well the four strains fared in a simple biome, and to see which parts of their genes made them more or less robust—to accomplish this feat they created a very large number of mutant strains (87,000 to 167,000) of the four species and set them loose in the mouse gut and then monitored them to see how well they fared by noting how many samples were found at given time intervals.
Then, to gain a better understanding of how a biome adapts to a change in diet, the mice were divided into four groups, two that were put on varying diets that swung from high fat and sugar to low fat and high complex carbohydrates and two that were put on non-varying diets to serve as a control. The researchers noted that the mice fed the varying diets developed biomes that were more complex than those with bland diets. The group was also were able to spot 15 genes that could be linked to amino acid metabolism and found that the simple gut biome they had created responded to a polysaccharide that could possibly serve as a probiotic.
More information: Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides, Science 2 October 2015: Vol. 350 no. 6256. DOI: 10.1126/science.aac5992
ABSTRACT
Libraries of tens of thousands of transposon mutants generated from each of four human gut Bacteroides strains, two representing the same species, were introduced simultaneously into gnotobiotic mice together with 11 other wild-type strains to generate a 15-member artificial human gut microbiota. Mice received one of two distinct diets monotonously, or both in different ordered sequences. Quantifying the abundance of mutants in different diet contexts allowed gene-level characterization of fitness determinants, niche, stability, and resilience and yielded a prebiotic (arabinoxylan) that allowed targeted manipulation of the community. The approach described is generalizable and should be useful for defining mechanisms critical for sustaining and/or approaches for deliberately reconfiguring the highly adaptive and durable relationship between the human gut microbiota and host in ways that promote wellness.
© 2015 Medical Xpress