Computer simulations explore how Alzheimer's disease starts

A new Rice University study uses computer simulations to explore the initial steps of the molecular process that leads to Alzheimer's disease.
The disease starts by the aggregation of a common protein called amyloid beta. The Rice study is the first to model the energy landscape of the assembly of many copies of the pathogenic protein into its toxic form.
The research led by Professor Peter Wolynes of Rice's Center for Theoretical Biological Physics is detailed this week in the Proceedings of the National Academy of Sciences.
Wolynes and his team are pioneers in the development of the energy landscape theory for proteins. All proteins start as chains of amino acids with their sequence determined by DNA, and each arrangement of the chain has a particular energy associated with it. The map of these energies and structures is called the energy landscape.
For most proteins, the energy landscape guides the protein to fold into its useful functional shape. Wolynes and his colleagues, developed a computer program, called AWSEM (associative memory, water mediated, structure and energy model), that simulates the process and can predict the most important functional structures.
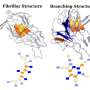
A single molecule of the amyloid beta protein by itself does not fold into a definite structure and also appears to be harmless, Wolynes said. The disease starts when several molecules come together to form an oligomer. The structures of such oligomers, which are thought to be the toxic agents, has been unknown, he said.
At a later stage, the oligomers change into fibrillar plaques that appear to be less toxic.
The Rice researchers used AWSEM to analyze oligomers of up to eight monomers in the prefibrillar form to see how they change into fibers.
In the oligomer made of six monomers, for instance, they found that this transition takes place when self-recognizing sequences on separate protein molecules interacted with each other rather than with their prefibrillar partners. These structures, they wrote, then form the stable core of a propagating amyloid aggregate.
"A few years ago, there were experiments on the misfolding of a beta protein called titin, and we started to study its aggregation," Wolynes said. "We discovered this interesting thing, that it wasn't the whole protein but only particular parts of the protein that were responsible for misfolding in amyloidogenic regions we found.
"It turned out there were very small lengths of about five amino acids that were extra sticky to each other," he said. "The surprise was that while the overall molecule was foldable, there were five to 10 residues that, if you had another copy of the same molecule, would be able to stick together and cause aggregation."
One problem with previous studies was that the protein concentration changes as the aggregation process proceeds, Wolynes said. An important mathematical development using energy landscape theory allowed the Rice researchers to correct for that effect.
With that correction, the simulation of oligomers of various complexity predicted solubilities that matched across the board what had been seen in experiments, said Weihua Zheng, a Rice research scientist and lead author of the study.
"One of the reasons you might get Alzheimer's is that you overproduce this protein," Zheng said. "That's why solubility is a key number: You want amyloid beta to remain soluble."
The problem may lie in the production of enzymes that keep oligomer concentrations in check, Wolynes said. Inhibiting such enzymes is being explored as a route for treatment with drugs, he said.
The simulations also demonstrated that the larger the oligomer, the greater the potential for forming fibrils, Wolynes said. Smaller oligomers of three or four monomers were predicted by the simulations to form cylindrins, barrel-shaped structures that other researchers have suggested could drill into and damage cell membranes. "In future work, we want to look at whether these cylindrical structures actually could penetrate a membrane in simulations," he said.
The researchers also noticed that just at the point of conversion, oligomers slightly unfold before going fibrillar. "We call that backtracking," he said. "It may explain a couple of things, like why these oligomers accumulate rather than all converting to the more benign fibrils."
In addition to enabling examination of the mechanism of amyloid beta aggregation, the simulations let the lab analyze the effects of point mutations—including hereditary variants called Dutch and Arctic—that are associated with early onset Alzheimer's. In both cases, the mutations seemed to make aggregation faster by exposing hydrophobic segments, though a third mutation, called Iowa, "seems to be mildly protective," Wolynes said.
The team will next simulate aggregation from amyloid beta proteins with 42 amino acids, for which structures of the fibers have recently been revealed. "It's thought that these aggregate more readily than the 40s we studied here, and we hope to see why these are different—or not," he said.
More information: Exploring the aggregation free energy landscape of the amyloid-β protein (1–40), www.pnas.org/cgi/doi/10.1073/pnas.1612362113