'Mysterious' non-protein-coding RNAs play important roles in gene expression
In cells, DNA is transcribed into RNAs that provide the molecular recipe for cells to make proteins. Most of the genome is transcribed into RNA, but only a small proportion of RNAs are actually from the protein-coding regions of the genome.
"Why are the non-coding regions transcribed at all? Their function has been mysterious," said Shelley Berger, PhD, a professor of Cell and Developmental Biology and director of the Penn Epigenetics Institute in the Perelman School of Medicine at the University of Pennsylvania.
Berger and Daniel Bose, PhD, a postdoctoral fellow in her lab, study the regulation of gene expression from enhancers, non-coding regions of the genome more distant from protein-coding regions. Enhancers boost the rate of gene expression from nearby protein-coding genes so a cell can pump out more of a needed protein molecule. A mysterious subset of non-coding RNAs called enhancer RNAs (eRNAs) are transcribed from enhancer sequences. While these are important for boosting gene expression, how they achieve this has been completely unknown.
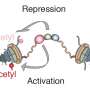
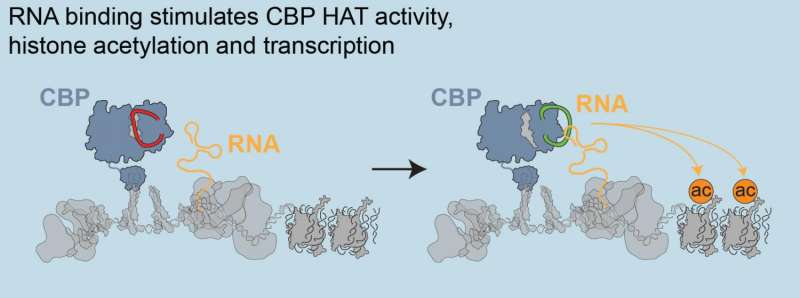
Shedding new light on these elusive eRNAs, they showed that CBP, an enzyme that activates transcription from enhancers, binds directly to eRNAs. This simple act controls patterns of gene expression in organisms by regulating acetylation, a chemical mark that directs DNA tightly packed in the nucleus of cells to loosen to promote transcription. Their findings are published this week in Cell.
"The cells in our bodies share the same genes and DNA sequences, and differ only in how these genes are expressed," Bose said. "Enhancers and eRNAs are critical for this process. Our work shows an exciting new way that eRNAs produce these different patterns of gene expression. We asked if eRNAs work directly with CBP, and found that they do."
Using biochemical assays, they showed that the region of CBP that binds to RNA also can regulate the ability of CBP to work with chemical mark. By binding to this region, eRNAs can directly stimulate CBPs' acetylation activity.
"There is increased interest in enhancers and eRNAs in the cancer biology world because defective enhancers can cause too much or too little of a protein to be made, or can cause the coding region to be turned off or on, or can make a protein at the wrong time," Berger said. Knowing more about how enhancers and eRNAs function will help oncologists, since recent DNA sequencing of tumors from humans show that numerous mutations associated with cancers and other diseases occur in enhancer regions of the genome—not in protein-coding regions.
"Fundamentally, this is important science because we show that enhancer RNAs have a key role throughout the genome and body to guide protein production," Berger said. "We identified, across the genome, that enhancer RNAs were the most common type of RNA that bound to CBP, and that by making this interaction, eRNAs play a crucial role in regulating CBP activity and gene expression."