Disease-associated genes routinely missed in some genetic studies
Whole-exome DNA sequencing—a technology that saves time and money by sequencing only protein-coding regions and not the entire genome—may routinely miss detecting some genetic variations associated with disease, according to Penn State researchers who have developed new ways to identify such omissions.
Whole-exome sequencing has been used in many studies to identify genes associated with disease, and by clinical labs to diagnose patients with genetic disorders. However, the new research shows that these studies may routinely miss mutations in a subset of disease-causing genes—associated with leukemia, psoriasis, heart failure and others—that occur in regions of the genome that are read less often by the cost-saving technology. A paper describing the research appeared online April 13 in the journal Scientific Reports.
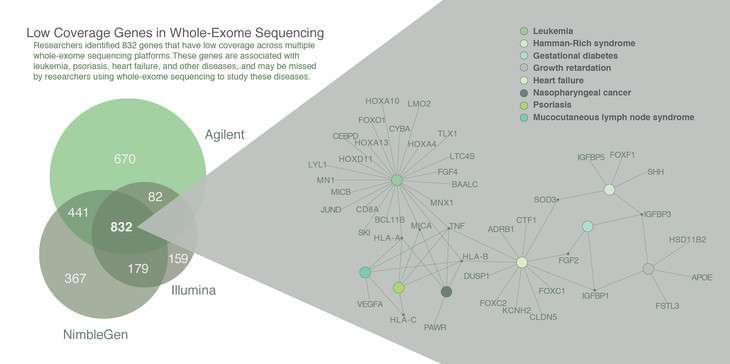
"Although it was known that coverage—the average number of times a given piece of DNA is read during sequencing—could be uneven in whole-exome sequencing, our new methods are the first to really quantify this," said Santhosh Girirajan, assistant professor of biochemistry and molecular biology and of anthropology at Penn State and an author of the paper. "Adequate coverage—often as many as 70 or more reads for each piece of DNA—increases our confidence that the sequence is accurate, and without it, it is nearly impossible to make confident predictions about the relationship between a mutation in a gene and a disease. In our study, we found 832 genes that have systematically low coverage across three different sequencing platforms, meaning that these genes would be missed in disease studies."
The researchers developed two different methods to identify low-coverage regions in whole-exome sequence data. The first method identifies regions with inconsistent coverage compared to other regions in the genome from multiple samples. The second method calculates the number of low-coverage regions among different samples in the same study. They have packaged both methods into an open-source software for other researchers to use.
"Even when the average coverage in a whole-exome sequencing study was high, some regions appeared to have systematically low-coverage," said Qingyu Wang, a graduate student at Penn State at the time of the research and the first author of the paper.
Low-coverage regions may result from limited precision in whole-exome sequencing technologies due to certain genomic features. Highly-repetitive stretches of DNA—regions of the genome where the same simple sequence of As, Ts, Cs and Gs can be repeated many times—can prevent the sequencer from reading the DNA properly. Indeed, the study showed that at least 60 percent of low-coverage genes occur near DNA repeats. As an example, the gene MAST4 contains a repeated sequence element that leads to a three-fold reduction in coverage compared to non-repeating sequences. Even when other genes have sufficient coverage, this region of the MAST4 gene falls well below the recommended coverage to detect genetic variations in these studies.
"One solution to this problem is for researchers to use whole-genome sequencing, which examines all base pairs of DNA instead of just the regions that contain genes," said Girirajan. "Our study found that whole-genome data had significantly fewer low-coverage genes than whole-exome data, and its coverage is more uniformly distributed across all parts of the genome. However, the costs of whole-exome sequencing are still significantly lower than whole-genome sequencing. Until the costs of whole-genome sequencing is no longer a barrier, human genetics researchers should be aware of these limitations in whole-exome sequencing technologies."
More information: Qingyu Wang et al. Novel metrics to measure coverage in whole exome sequencing datasets reveal local and global non-uniformity, Scientific Reports (2017). DOI: 10.1038/s41598-017-01005-x