Scientists characterize regulatory DNA sequences responsible for human diseases
Scientists from the Children's Medical Center Research Institute at UT Southwestern (CRI) have developed an innovative system to identify and characterize the molecular components that control the activities of regulatory DNA sequences in the human genome.
The genome, which is the complete complement of human DNA, including all protein-coding genes, has nearly 3 billion base pairs. Despite its vast size, only 2 percent of our genome encodes for proteins. The other 98 percent is comprised of noncoding regions that regulate where and when the protein-coding genes are activated. These noncoding regions have repeatedly been identified by human genetics and cancer genomic studies as potential drivers for human diseases such as cancer.
A better understanding of these regulatory regions and the underlying principles that guide when genes are turned on and off is necessary to uncover how diseases develop and to find new treatments. However, the tools to identify these noncoding regions and to understand how they work are limited. They require the prior identification of the protein factors that regulate these regions, depend on the availability of reagents such as antibodies, and often need sophisticated genetic manipulations.
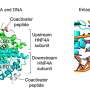
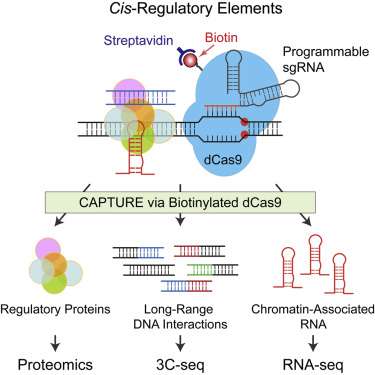
The new system, developed by researchers in the Dr. Jian Xu lab and published in the latest issue of Cell, is paving the way for an in-depth look at these regulatory genetic elements. This system, named CAPTURE (CRISPR Affinity Purification in situ of Regulatory Elements), provides an approach to simultaneously isolate genomic sequence-associated proteins as well as their RNA and DNA interactions.
"The ability that CAPTURE gives us to isolate and analyze the entire set of factors that regulate our DNA offers many possibilities to study how different proteins control genome function in cancer and stem cells," said Dr. Xu, senior author of the study and Assistant Professor in CRI at UTSW and the Department of Pediatrics. "It also opens up a completely new avenue to find new drug targets."
The CAPTURE method was developed by repurposing the CRISPR genomic editing system, including the CRISPR associated protein 9 (Cas9) - an RNA-guided enzyme that binds to DNA. CAPTURE works by using guide RNAs to direct a deactivated version of Cas9 (dCas9) to the DNA elements that researchers want to study. Then, dCas9 - along with other proteins, RNA, and DNA sequences associated with dCas9's position on the chromosome (its genomic loci) - can be isolated and studied. This makes it possible to identify and characterize genomic regulatory regions, and their associated proteins, throughout the genome.
Using CAPTURE, Dr. Xu's laboratory successfully identified many known and new human telomere-associated proteins as a proof of principle. Telomeres, which are short, repetitive DNA sequences on the ends of chromosomes, protect our chromosomes from fraying or fusing with neighboring chromosomes. Next, researchers uncovered new mechanisms regulating aberrant beta-globin gene expression in human blood cells. Beta-globin is a vital part of a larger protein known as hemoglobin that is responsible for the exchange of oxygen and carbon dioxide between our lungs and body tissues. Altered expression of beta-globin genes is associated with inherited hemoglobin disorders, such as sickle cell disease, currently affecting 5 percent of the world's population.
"The unbiased analysis of the genome by CAPTURE provides biomedical researchers with a powerful new tool to decipher underlying regulatory principles. This new tool will advance our understanding of the human genome and genetic variations in a variety of diseases," Dr. Xu said.
More information: In Situ Capture of Chromatin Interactions by Biotinylated dCas9, Cell (2017). DOI: 10.1016/j.cell.2017.08.003 , www.cell.com/cell/fulltext/S0092-8674(17)30891-7