These mutations could be key to understanding how some harmful conditions develop
A team of researchers led by a bioinformatician at the University of California San Diego has developed a method to help determine whether certain hard-to-study mutations in the human genome, called short tandem repeats or microsatellites, are likely to be involved in harmful conditions.
The team, which also includes scientists from the New York Genome Center, Harvard University, and the Massachusetts Institute of Technology, details their findings in the Sept. 11 issue of Nature Genetics.
In short tandem repeats, sequences of one to six of DNA's basic components, called nucleotides, repeat over and over again, sometimes up to hundreds or thousands of times.
These mutations already have been implicated in about 30 conditions. The best known is perhaps Huntington's Disease, which causes the progressive breakdown of nerve cells in the brain. About 30,000 people suffer from the condition in the United States. These people all have more than 40 copies of a specific repeat. The more copies they have, the sooner they are affected by the disease and the more severe it is.
The Nature Genetics paper is part of the ongoing, decades-long effort to pinpoint harmful mutations in the human genome. Tandem repeats are often overlooked in these efforts, and have sometimes been disregarded as "junk DNA." But researchers led by Melissa Gymrek, an assistant professor at UC San Diego, believe that tandem repeats are likely to play key roles in human health and need to be studied in depth.
"When you look for signals for disease in the human genome, you get too many answers. We are looking for a way to narrow these answers down," said Gymrek, who holds appointments at both the UC San Diego School of Medicine and the Jacobs School of Engineering.
In the next step of their research, scientists plan to use their model to examine the genomes of families with autistic members.
Analyzing repeats
Tandem repeats are difficult to analyze with current genome sequencing techniques. That's because they're usually fairly long, and current tools usually look only at short pieces of DNA. In addition, the process of amplifying DNA for sequencing creates more errors that get in the way.
In this paper, researchers detail how they were able to create a mathematical model that predicts how frequently and in what way the repeats appear and mutate in the human genome. Gymrek and colleagues were able to do this because of the extraordinary amount of genetic data that they had access to—more than 1.5 million repeats from the genomes of 300 individuals.
The researchers based their new algorithm on a method called MUTEA that they previously developed to precisely estimate individual mutation rates for tandem repeats on the Y chromosome. They modified the algorithm so it would analyze pairs of DNA variations, called haplotypes. The key insight the method provided is that different classes of mutations happen at regular, predictable intervals in time, constituting what they refer to as a molecular clock. This clock can be used to determine how often mutations occur within a genome.
Finding constraints
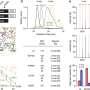
Next, the researchers used the model to calculate actual mutation rates and compare those to expected mutation rates. This is what geneticists call constraint. For example, regions of the genome that are home to mutations that occur early in life and lead to severe health conditions tend to have fewer mutations in the population than expected by chance—geneticists say they're highly constrained. That's because those suffering from these conditions, like autism, are less likely to pass their genes on to the next generation. Regions of the genome that cause diseases that occur later in life, after patients have had children, like Huntington's Disease, are usually not constrained.
The team used their model on a number of different tandem repeats related to both late and early onset conditions, such as limb malformations. The model correctly identified that repeats involved in early-onset conditions were subject to constraint. They calibrated their method by using a set of tandem repeats that are not associated with specific conditions, which the FBI uses to identify people. As expected, these repeats mutate at the expected rate and are not constrained.
Gymrek and her team are now getting ready to apply their model to find signals for other conditions inside the human genome.
More information: Melissa Gymrek et al. Interpreting short tandem repeat variations in humans using mutational constraint, Nature Genetics (2017). DOI: 10.1038/ng.3952