Computer simulations identify chemical key to diabetes drug alternatives

Jeremy Smith, Governor's Chair for Molecular Biophysics at the University of Tennessee, Knoxville, and director of the Center for Molecular Biophysics at Oak Ridge National Laboratory, has worked with a research team from the UT Health Science Center to discover a chemical compound that could lower sugar levels as effectively as the diabetes drug metformin but with a lower dose.
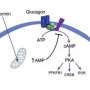
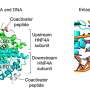
Smith, together with Jerome Baudry, of the University of Alabama at Huntsville, and graduate student Karan Kapoor, of the University of Illinois at Urbana-Champaign, used high-performance computing to create sophisticated simulations to suggest chemicals that could activate GPRC6A, a protein that regulates sugar levels by simultaneously correcting abnormalities in pancreatic insulin secretion, glucose uptake into skeletal muscle, and liver regulation of glucose and fat metabolism. The UTHSC team verified its potency and used this starting point to design an even more effective chemical.
"This chemical compound lowers sugar levels in mice as effectively as metformin, but with a 30-times lower dose," Smith said. "It therefore is a good starting point for the development of a new and effective drug to fight diabetes."
This new approach to diabetes drug discovery has been published in PLOS One, a peer-reviewed open access scientific journal.
Leading the UTHSC research team is Darryl Quarles, UT Medical Group Endowed Professor of Nephrology, director of the Division of Nephrology, and associate dean for Research in the College of Medicine at UTHSC.
With more than 400 million people suffering from Type 2 diabetes worldwide, the global cost of medicine and prevention is close to a trillion dollars annually. Metformin, a drug that lowers the liver's production of sugar and decreases risk of mortality, is currently recommended as a first-line treatment. However, there is a need for alternative treatment options when patients are not responsive to metformin.
Smith's computations found several chemicals that might activate the protein, and the Quarles laboratory at UTHSC tested each. A team of UTHSC medicinal chemists then used the results to synthesize related molecules for pre-clinical testing, and a chemical called DJ-V-159 was found to be highly potent in stimulating insulin secretion and lowering sugar levels in mice.
More information: Min Pi et al. Computationally identified novel agonists for GPRC6A, PLOS ONE (2018). DOI: 10.1371/journal.pone.0195980