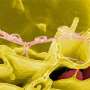
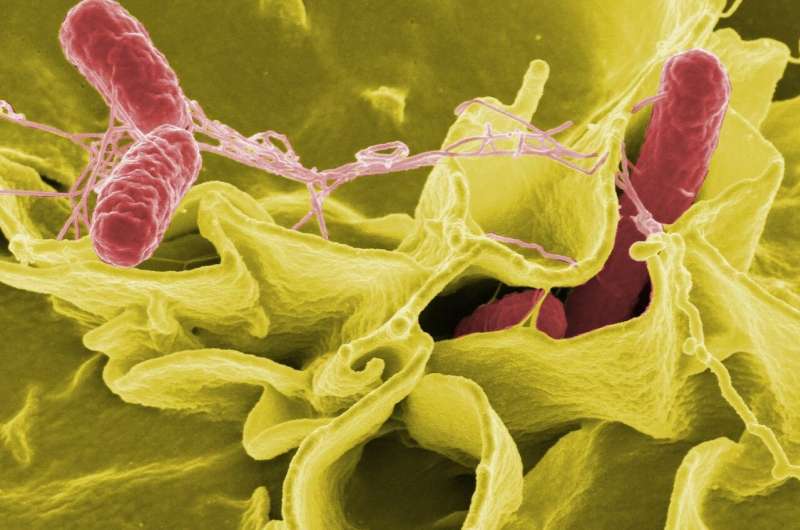
Salmonella, gene swapping and antibiotic resistance: Five questions with Sid Thakur
Sid Thakur is a professor of population health and pathobiology and the director of NC State's and the College of Veterinary Medicine's Global Health programs. He studies antibiotic resistance in Salmonella and how antibiotic resistant Salmonella affect food animal and human populations. In his latest work, Thakur compared the genomes of over 200 different strains of Salmonella, looking for genetic similarities across strains. He spoke with the Abstract about what this work can tell us, and how it may help to keep us and our food supply safe.
The Abstract (TA): In this paper you compared the genomes of 200 different Salmonella strains taken from humans, pigs, chickens and farm environments. Are there more varieties of Salmonella out there?
Thakur: Salmonella has more than 2,500 different serotypes. The common ones we typically encounter during outbreaks include Typhimurum, Heidelberg, Enteritidis, Newport, 4[5], 12:i:- and Muenchen, to name a few. Many of these serotypes are host adapted, meaning they predominantly appear in one host – for example, Enteritidis is usually found in eggs. Many others exist in multiple hosts – Typhimurium occurs in humans, animals and the environment.
TA: Why did you choose the strains you chose?
Thakur: The main reason is because these strains were isolated from multiple sources including humans, animals and the environment. These strains are responsible for many of the recent outbreaks in humans, and we wanted to determine if these Salmonella serotypes from multiple sources had similar or diverse characteristics. Finally, many of these strains are resistant to multiple antimicrobials, and it is important to compare their genomic profiles so we can understand what makes them resistant.
TA: What can comparing the DNA of different Salmonella strains tell us?
Thakur: Genome comparison can reveal a lot of information that other molecular-based approaches like PCR lack. We can identify genomic regions that are similar or different. We can get insights into the drug resistance and virulence profiles across multiple genomes and understand how the strains are evolving under different selective pressures. Genome comparison can help us understand whether different strains are exchanging genomic material, identify new emerging pathogenic strains and compare the profiles at the global level.
TA: Different Salmonella strains can swap genes. Why do they do this? Is there a way to prevent it?
Thakur: Pathogens are continuously evolving trying to counteract the harmful environment they encounter. The Darwinian principle "survival of the fittest" holds true here. If pathogens don't evolve then they will not survive. Salmonella is a good example and specific serotypes of this pathogen are more diverse at the genomic level. One good example is Salmonella Typhimurium, which is a diverse serotype and hence has the ability to survive in multiple hosts. Flexibility in the genomes allows pathogens to make changes in their DNA and, in many cases, exchange genes with other pathogens. As such, Typhimurium is responsible for many outbreaks in humans that get exposed to it through different hosts, including food products and the environment. The swapping of genes and genomic material is a continuous process and difficult to control or prevent. That is what makes this field so challenging and exciting to work in at the same time.
TA: What was the most interesting result of the work? Were there any surprises?
Thakur: We detected close relationships between different Salmonella serotypes across multiple hosts, potentially indicating zoonotic transmission, where the infection is transmitted from an animal to a human. We also compared different approaches to improve the resolution of Salmonella phylogenetic trees that we create using the genomic data. We identified the same drug resistance genes, plasmid types and virulence genes distributed across multiple hosts – and the number of hosts and extent of the switching was surprising and interesting to study.