April 17, 2020 report
Single cell division error triggers cascade of mutations that generate features of cancer
A team of researchers affiliated with multiple institutions in the U.S. and the U.K. has found that a single error during cell division can trigger a cascade of mutational events, generating many of the features found in cancer genomes. In their paper published in the journal Science, the group describes using live cell imaging and single-cell whole-genome sequencing to study the repercussions of single-cell mutational events on downstream cellular development. Jacob Paiano and André Nussenzweig with the National Cancer Institute at the National Institutes of Health have published a Perspective piece in the same journal issue outlining some of the recent history behind chromothripsis research and the work done by the researchers with this new effort.
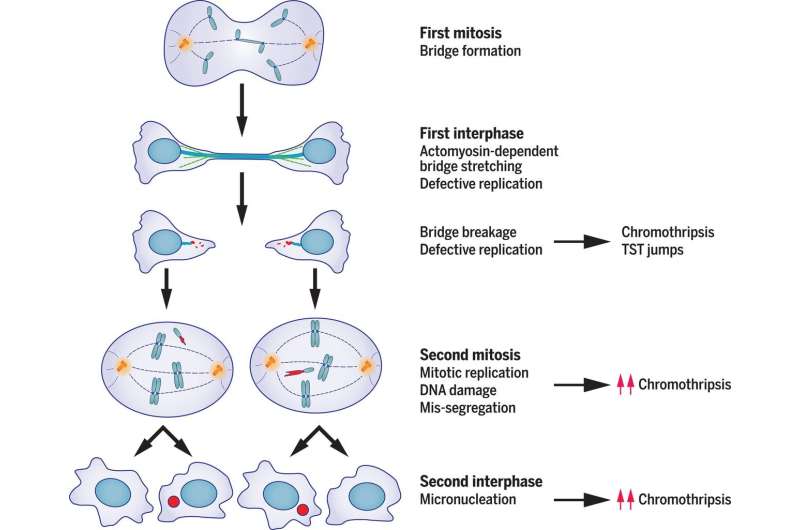
Over the past several generations, medical researchers have assumed that the complexity of the genomes of many cancer tumors arises gradually—small mutations during replication added together over multiple generations can result in a large mix of errors. More recently, researchers have found that cancer genomes can evolve more quickly than previously believed due to a single catastrophic mutation. They have also found that the type of mutations seen in tumors can come about quickly due to a wide variety of processes, most particularly those occurring during the chromosome breakage-fusion-bridge cycle and chromothripsis (a process in which thousands of chromosomal rearrangements occur due to a single event). In this new effort, the researchers sought to find out whether the different processes that lead to such tumors might be related.
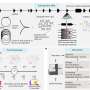
To learn more, the team used an approach that involved live-cell imaging and single-cell whole-genome sequencing—this technique allowed them to track both the cellular and genetic repercussions that ensued after chromatin bridge generation—which connected daughter cells. They found that a one-off dicentric chromosome fusion event could trigger a cascade of mutations in subsequent daughter cell divisions, which could lead to instability in the genome and mutational heterogeneity in the genes of the cells that followed. This finding provides some insight into the etiology of cancer, which could lead to new research into preventing mutations that are associated with chromothripsis.
More information: Neil T. Umbreit et al. Mechanisms generating cancer genome complexity from a single cell division error, Science (2020). DOI: 10.1126/science.aba0712
© 2020 Science X Network