Researcher identifies gene teams working in subregions of brain
You must first understand how something works normally before you can figure out why it's broken.
That's the idea behind brain research by Yuqing "Iris" Hang, a Clemson University graduate student pursuing a doctorate in biochemistry in the College of Science's Department of Genetics and Biochemistry.
Hang is studying how genes interact in a normal brain.
"When studying a disease trait, we always need to compare that trait to the normal condition," she said.
An article published recently in Scientific Reports titled "Exploration into biomarker potential of region-specific brain gene co-expression networks," describes Hang's research.
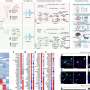
Hang used RNA-seq gene expression profiles from the National Institutes of Health Genotype-Tissue Expression (GTEx) project and software called the Knowledge Independent Network Construction (KINC) to sort thousands of genes into gene groups that team up in specific parts of the normal human brain. Clemson professor Alex Feltus and former graduate student Stephen Ficklin, who is now an assistant professor at Washington State University, developed KINC. Hang is a student in Feltus' Systems Genetics Lab.
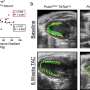
Hang then constructed six mini gene co-expression networks for a normally functioning brain and conducted additional statistical tests to reveal correlations between genes and cellular pathways in the brain. In a gene co-expression network, two genes with a high likelihood of interacting with each other will be connected by a line called an edge.
During her analysis, Hang found teams of genes working in all parts of the brain—but only in the brain—and she also found teams of genes working in subregions of the brain.
"We have brain biomarkers—genes that are co-expressed pair-wise in different brain tissues—that are evidence that they are probably doing something normal in those tissues they're not doing in other tissues. They have some collective function," Feltus said. Researchers can now test each of those gene teams to see if gene pairs are changing in brain tumors or in people with intellectual disabilities.
"Now we know what genes are interacting normally in the brain, and then we can test and see if those interactions don't exist in disease," he said.
One notable finding is that genes in the six brain mini gene co-expression networks showed higher mutation rates in tumors than in matched sets of random genes.
"This means those genes could play an important role in brain development. The mutation of those genes would increase the chances of getting a brain tumor or brain-related disease," Hang said.
Feltus called Hang's research a step toward the goal of finding gene teams that are controlling phenotypes.
"One hundred years from now, I don't think there will be any cancer, and I think we'll understand how intellectual disabilities are occurring," he said.
More information: Hang, Y., Aburidi, M., Husain, B. et al. Exploration into biomarker potential of region-specific brain gene co-expression networks. Sci Rep 10, 17089 (2020). doi.org/10.1038/s41598-020-73611-1