This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
DNA region that directs development of craniofacial structures identified
Craniofacial birth defects, including cleft lip and palate, are among the most common human congenital malformations. Now, FMI researchers have identified a DNA region containing multiple regulatory elements that interact with genes across distant chromosomal neighborhoods, ensuring that specific facial structures develop in the right place. The findings may provide new insights into normal head development as well as craniofacial birth defects.
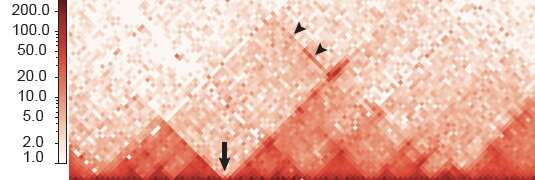
Craniofacial anomalies occur because of defects in neural crest cells, which give rise to the face and skull. Working in mouse neural crest cells, researchers led by Filippo Rijli identified 2,232 "super-enhancers"—clusters of DNA regions that control how genes are expressed. More than 145 of these super-enhancers targeted genes involved in establishing neural crest cell "positional" identity, which is connected to the cells' location in the face as it develops. The activity of these super-enhancers and their target genes thus ensures that the right facial structure is built in the right place.
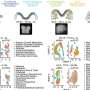
In a specific group of neural crest cells, the team identified a stretch of DNA that contains multiple super-enhancers and is partitioned into two regions, which the researchers called HIRE1 and HIRE2. Both are located in the same topologically associating domain, or TAD. TADs are chromosomal neighborhoods in which DNA sequences are more likely to interact with one another than with those in other TADs. But Rijli's team found that HIRE1 and HIRE2 interact with two genes called Hoxa2 and Hoxa3, which are located in a distant TAD. These genes are required for the formation of certain ear and neck structures.
Deleting HIRE2 dampened Hoxa2 levels, resulting in microtia—a congenital facial deformity where the external ear does not fully develop. Instead, HIRE1 deletion resulted in severe external and middle ear abnormalities as well as defects in neck structures associated with the loss of both Hoxa2 and Hoxa3 expression.
Published in Nature Communications, the findings suggest that mutations in regulatory regions that "create bridges" to contact target genes in distant chromosomal neighborhoods may result in craniofacial birth defects that have not been linked to specific genes, the researchers say.
More information: Sandra Kessler et al, A multiple super-enhancer region establishes inter-TAD interactions and controls Hoxa function in cranial neural crest, Nature Communications (2023). DOI: 10.1038/s41467-023-38953-0