August 7, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Unbiased omics method finds in vivo host restriction factors for HIV-1
Research led by the Walter Reed Army Institute of Research, Maryland, has confirmed previous research findings of host restriction factors that target HIV-1. In a paper, "Single-cell transcriptomics identifies prothymosin α restriction of HIV-1 in vivo," published in Science Translational Medicine, the authors detail how omics can uncover correlations in the search for cure strategies.
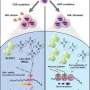
Single-cell transcriptomics was used to identify proteins active in the cells of 14 patients with HIV-1 infection. The team narrowed down a potential list of antiviral proteins called host restriction factors by correlating the patient gene expression with viral RNA (vRNA) quantities within individual cells. These proteins, part of the body's innate immune system, recognize and interfere with specific steps of the replication cycle of viruses, thereby blocking infection.
The team also compared the sequencing of a participant with the highest plasma viral load, which revealed vRNA transcription to be inversely correlated with the expression of the gene PTMA, which encodes prothymosin αlpha (ProTα).
This association was then validated in 28 additional participants outside the initial study. Overexpression of prothymosin α in vitro confirmed that this cellular factor inhibits HIV-1 transcription and infectious virus production.
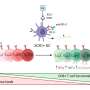
The researchers found that HIV-1 RNA (vRNA) in individual immune cells correlates with specific clinical measurements. Using single-cell analysis, researchers identified cells with vRNA from participants during Acute HIV Infection (AHI) and antiretroviral therapy (ART) stages.
The vRNA+ cells were mostly found within CD4+ T cell subsets. Correlation analysis revealed that the frequency of vRNA+ CD4+ T cells was positively associated with measures such as total cell-associated HIV-1 DNA and plasma viral load. This suggests that the presence of vRNA in these cells is biologically significant and relates to HIV-1 clinical parameters.
In a confirmation of the method, the ProTα HIV-1 restriction highlighted was previously discovered 17 years ago by researchers at Mount Sinai School of Medicine, New York, and described in a paper, "Novel Function of Prothymosin Alpha as a Potent Inhibitor of Human Immunodeficiency Virus Type 1 Gene Expression in Primary Macrophages," published in the Journal of Virology.
The previous research also observed infection of human T cells with HIV-1 associated with reduced detectable amounts of ProTα mRNA. The Mount Sinai study found ProTα activity significantly suppressed HIV-1 replication in primary macrophages in a dose-dependent manner but had little to no effect on HIV-1 in CD4+ T cells.
The current study demonstrates the use of unbiased omics technologies to identify potential antiviral factors like ProTα for HIV-1 and other viral prevention and cure strategies.
More information: Aviva Geretz et al, Single-cell transcriptomics identifies prothymosin α restriction of HIV-1 in vivo, Science Translational Medicine (2023). DOI: 10.1126/scitranslmed.adg0873
© 2023 Science X Network