This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
17-gene signature linked to remission after triple-negative breast cancer treatment

Mayo Clinic researchers have discovered a distinctive pattern in a specific set of 17 genes that may be associated with remission after treatment for triple-negative breast cancer. The multi-omics study, published in Breast Cancer Research, highlights the potential for further investigating this signature as a target for individualized medicine.
Approximately 10–15% of breast cancers fall into the category of "triple-negative," indicating that their growth is not driven through the hormone receptors for estrogen or progesterone, nor by the human epidermal growth factor receptor 2 (HER2). In contrast, other breast cancers have at least one of these receptors, allowing for treatments that can target or block them to inhibit the cancer's progression.
Triple-negative breast cancer is known for its resistance to targeted therapies, leaving chemotherapy as the primary treatment option. While most patients initially respond well to the standard neoadjuvant chemotherapy, nearly 20% of patients will experience recurrence after treatment.
"It's crucial that we create tools that assist clinicians in making informed treatment decisions," says Krishna Rani Kalari, Ph.D., a lead author of the study and an associate professor of biomedical informatics at the Center for Individualized Medicine. "This 17-gene signature is one of the candidate tools that shows potential. However, for this signature to have clinical utility, there will need to be studies evaluating whether alternative drugs/drug combinations may work in this group of patients."
Decoding pattern in 17 genes
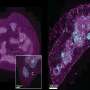
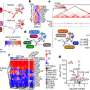
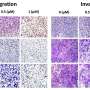
In their study, the researchers investigated tumor samples before and after treatment of patients with triple-negative breast cancer who failed to respond to neoadjuvant chemotherapy. Using multi-omics analysis and machine learning technologies, they examined changes in gene expression to understand the molecular underpinnings driving tumor recurrence.
Multi-omics is an emerging multidisciplinary field of biological sciences that encompasses genomics, proteomics, epigenomics, transcriptomics, metabolomics and more.
The team identified 17 genes associated with relapse-free survival and found that many of the genes may play a role in triggering inflammation to initiate an immune system response. Dr. Kalari says failure to initiate immune response prevents the clearance of weakened tumor cells, thereby permitting the development of treatment resistance and disease re-occurrence.
Quest for individualized medicine
The study is an extension of Mayo Clinic's clinical prospective breast cancer study known as Breast Cancer Genome-Guided Therapy Study (BEAUTY). The goal of the study is to determine why some patient's tumors respond to chemotherapy while others don't, and to use that knowledge as a catalyst to develop personalized medicine therapies.
Next, the team plans to further investigate the gene signature and test therapeutic strategies.
More information: Xiaojia Tang et al, Integration of multiomics data shows down regulation of mismatch repair and tubulin pathways in triple-negative chemotherapy-resistant breast tumors, Breast Cancer Research (2023). DOI: 10.1186/s13058-023-01656-x