This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study evaluates protein that regulates pigment cell development for role in skin cancer
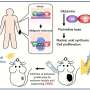
The microphthalmia-associated transcription factor (MITF) is the master regulator of pigment cell development and, as a lineage survival oncogene, plays a crucial role in the skin cancer melanoma and its resistance to therapy. How MITF distinguishes between its seemingly incompatible differentiation and proliferation-associated targets in the genome has been a bit of a puzzle.
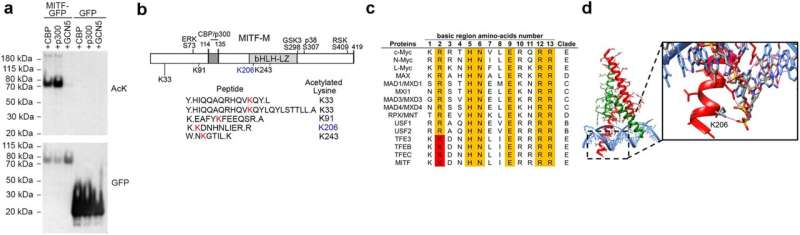
Ludwig Oxford's Pakavarin Louphrasitthiphol, Colin Goding and colleagues discovered that the ability of MITF to bind DNA is inhibited by CBP/p300-mediated acetylation of its lysine residue 206, which preferentially directs its binding away from DNA elements involved in differentiation. This suggests an explanation for why a mutation of that residue—K206Q—is associated with Waardenburg syndrome, which is often characterized by defects in pigmentation of hair, skin and eyes.
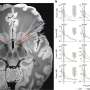
Reported in Nature Communications, the results also reveal that more than 40% of MITF molecules are tightly bound to DNA in the nucleus, with residence times of more than 100 seconds—compared to just a handful of seconds for most transcription factors. This makes MITF comparable to transcriptional repressor CTCF and polycomb repressive complex 1 (PRC1) and suggests that it might play similar roles in the establishment and maintenance of chromatin organization specific to the melanocyte lineage.
More information: Pakavarin Louphrasitthiphol et al, Acetylation reprograms MITF target selectivity and residence time, Nature Communications (2023). DOI: 10.1038/s41467-023-41793-7