This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
DNA sequencing reveals clonal evolution of tumor in childhood leukemia
Researchers at the RUDN Laboratory of Biology of Single Cells have studied the possibilities of DNA sequencing at the level of individual cells in patients with a rare form of childhood leukemia. The study revealed the mechanisms of the clonal evolution of the tumor and its transformation into a more aggressive form of leukemia at the level of individual subclones. It has also been shown that this technology can also be used as a very accurate tool for assessing the response of the disease to the treatment.
The results will help to use single-cell sequencing for the diagnosis and therapy selection in patients with this type of tumor. The results are published in npj Systems Biology and Applications.
Juvenile myelomonocytic leukemia (JMML) is a rare form of leukemia that develops in children in the first years of life. It occurs due to mutations in the RAS-signaling pathway in hematopoietic stem cells. As a result of these mutations, the regulation of hematopoiesis is disrupted, which leads to the excessive formation of some cells and a lack of others.
Without transplantation of donor stem cells (HSCT), such patients—in most cases—develop aggressive forms of leukemia that practically do not respond to therapy, or die from complications of the disease.
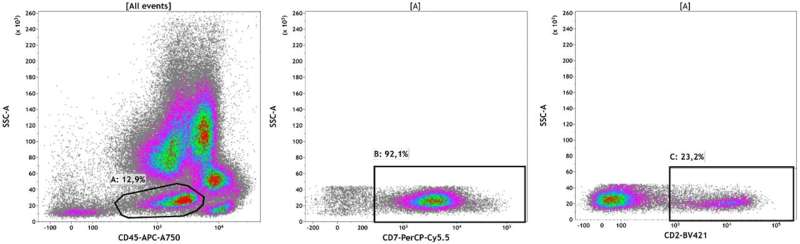
To diagnose and evaluate the prognosis, doctors need to know the nucleotide sequence of the studied genes and identify mutations in them. However, standard sequencing methods cannot always detect minor tumor subpopulations, which can subsequently lead to the progression of the disease. This limitation is because beam methods give averaged indicators and do not allow studying the characteristics of the tumor at the level of individual cells or subpopulations.
Scientists from RUDN University have reported on two cases of using single-cell DNA sequencing in patients with JMML.
"The method we used allowed us to find out the cause of the disease progression in therapy in two patients with JMML. It turned out that it was caused by the appearance of a new tumor clone with an additional mutation," said Egor Volchkov, intern researcher at the RUDN Research Institute of Molecular and Cellular Medicine.
"In addition, we were able to assess the level of chimerism, that is, the ratio between donor and recipient cells, after the HSCT, which was the basis for the appointment of immunotherapy, which allowed saving one of the patients. Such a thing would be practically impossible using standard sequencing methods, which give 'averaged' indicators, without reference to specific cells."
DNA sequencing of single cells transfers the study of the genome to the cellular level. It involves the isolation of individual cells in a fat emulsion, DNA barcoding, and the development of a library of the studied genome sites. The scientists used this method to analyze the blood of two patients with JMML in different periods of the disease. Pathology was detected in one of the patients at the age of three years, and in another at one month. In both of them, JMML led to acute myeloid leukemia.
When diagnosed, both patients had only one "classical" mutation in the RAS signaling pathway. Analysis of the material obtained during the transition of YUMML to acute myeloid leukemia showed the presence of clones with additional mutations, which most likely caused the progression of the disease. The DNA sequencing technology of single cells also allowed the scientists to measure the level of chimerism after HSCT, which is the main indicator of the effectiveness of transplantation and the basis for prescribing subsequent therapy.
"The ability to detect genetic changes among thousands of individual cells allows us to study the mechanisms of the development of MML at a fundamentally new level. The acquired knowledge already makes it possible to change the therapy in such patients and may become the 'gold standard' of diagnosis in such patients," said Volchkov.
More information: E. V. Volchkov et al, Potential value of high-throughput single-cell DNA sequencing of Juvenile myelomonocytic leukemia: report of two cases, npj Systems Biology and Applications (2023). DOI: 10.1038/s41540-023-00303-7