This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
A new gene classification system for gastric cancer
A research team has announced a new genetic classification system for gastric cancer by a multicenter study with the University of Texas MD Anderson Cancer Center. This research could lay the foundation for personalized gastric cancer treatment.
Five institutions including the MD Anderson Cancer Center, Korea University College of Medicine, CHA University Medical Center, Kyung Hee University School of Medicine, Yonsei University College of Medicine, and Sungkyunkwan University School of Medicine; participated in this multicenter study.
The research, published in the journal Molecular Cancer, was led by Professor Ju‑Seog Lee of MD Anderson Cancer Center and Professor Sang Cheul Oh of Division of Oncology/Hematology participated. Professor Sang‑Hee Kang of Department of Surgery and Professor Sun Young Yim of Division of Gastroenterology and Hepatology participated led as the co-lead authors.
Previously, the MD Anderson- Korea University research team also published the genetic classification system for liver cancer in Hepatology.
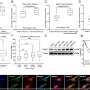
Gastric cancer tends to have genetic and clinical diversity. The research team analyzed the gene classification system for 8 gastric cancers that was published in the past and derived the result of 6 Consensus Genomic Subtypes (CGSs). This system classified gastric cancers from CGS1 to CGS6 depending on different gene expression patterns.
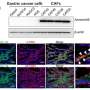
Each subtype had different characteristics and CGS1 showed the worst prognostic features. It had very high stem cell properties and low genetic modification. However, the analysis confirmed that CGS1 responded well to immunotherapy and treatments targeting IGF1R could be effective. CGS2 was enriched in typical epithelial cell gene expression. CGS3 and CGS4 showed high cloning number variation and had low responses to immunotherapy.
Nevertheless, CGS3 had a characteristic of HER2 gene activation and CGS4 had a characteristic of SALL4 gene activation. The team analyzed that treatments targeting those features would be effective. CGS5 had high mutation burden, which is a characteristic of microsatellite instability in tumors, and showed moderate response to immunotherapy. CGS6 was mostly positive for infectious mononucleosis (Epstein Barr) virus and had very high methylation levels. It showed a high response to immunotherapy.
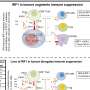
The research team not only classified genetics of gastric cancer, but also estimated the potential response rates of standard and experimental treatments (chemoradiotherapy, immunotherapy, etc.) for each subtype through systematic analysis of genome and proteome data. As a result, the CGS3 subtype showed especially great benefits in anticancer radiotherapy due to its iron-dependent cell death due to high level lipid peroxidation. Research suggested potential treatment subjects for each subtype.
Professor Sun Young Yim of the Division of Gastroenterology and Hepatology, one of the lead authors of this study, said, "Although the mortality rate of gastric cancer is decreasing due to the implementation of new therapies, it still is one of the major causes of death for cancer patients. We believe that this research is going to lay the foundation for personalized gastric cancer treatments."
More information: Yun Seong Jeong et al, Clinically conserved genomic subtypes of gastric adenocarcinoma, Molecular Cancer (2023). DOI: 10.1186/s12943-023-01796-w