This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers discover potential target for gastric cancers associated with Epstein-Barr virus
Scientists at The Wistar Institute have discovered a potential target for gastric cancers associated with Epstein-Barr Virus, and their study results are published in the journal mBio.
In the paper, Wistar's Tempera lab investigates the epigenetic characteristics of gastric cancer associated with the Epstein-Barr Virus: EBVaGC. In evaluating EBVaGC's epigenetics—the series of biological signals associated with the genome that determines whether a given gene is expressed—the Tempera lab highlights a target that could advance as a future treatment for this type of cancer.
The work of Italo Tempera, Ph.D., associate professor in the Gene Expression & Regulation Program in the Ellen and Ronald Caplan Cancer Center, at The Wistar Institute, and collaborators demonstrate that an epigenetically active compound called decitabine disrupts the genome of EBVaGC by epigenetically modifying the cancer's DNA, a finding that offers the potential for a new approach to treating EBVaGC.
"What we have identified is essentially a self-destruct button within this kind of cancer, and our paper shows that we figured out how to press that self-destruct button," said Tempera. "Normally, a latent virus that reactivates and starts to kill cells is a bad thing. But by switching that viral lytic process back on in these cancer cells by using epigenetic signaling, we're effectively getting the virus to kill the cancer cells that it's responsible for in the first place."
The research includes scientists from The Wistar Institute, The Coriell Institute for Medical Research, and Brigham and Women's Hospital of Harvard Medical School.
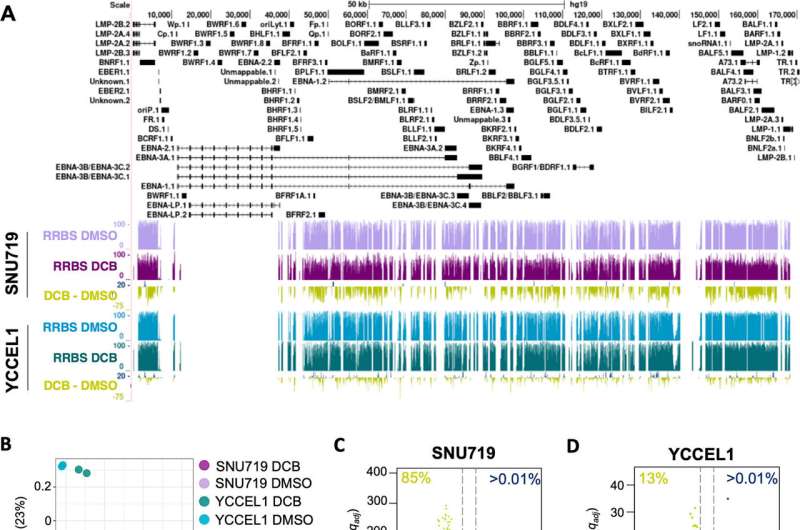
In EBVaGC, the cancer cells' DNA is hypermethylated: the DNA contains a high percentage of cytosine with a 5-methyl group attached to it (relative to normal, unmethylated cytosine). As a silencer of gene expression, DNA methylation allows EBV to remain latent. This methylation pattern plays a significant role in regulating the EBV latency-lysis cycle within the cancer cells.
DNA methylation, as an epigenetic factor, usually functions as a gene-silencing mechanism, particularly in certain regions of the genome; a methylated gene still exists within the genome—methylation does not delete the genetic information—but methylation can prevent the protein the gene encodes from being transcribed.
To disrupt this epigenetic profile, the researchers turned to decitabine, a compound known for its ability to reduce DNA methylation levels (i.e., to hypomethylate the DNA). Tempera and his co-authors treated two cell lines that were derived from EBVaGC tumors with decitabine. The cell lines that received the treatment demonstrated massive reductions in DNA methylation across the genome relative to the control as assessed by a variety of epigenetic assay techniques.
In observing the effects of decitabine treatment on EBVaGC, Tempera's team found a significant disruption of the cancer's epigenetic profile. The EBV genome within EBVaGC treated with decitabine resulted in widespread, mostly uniform hypomethylation of the EBVaGC epigenome (with a few regional exceptions).
Tempera and his co-authors discovered that the hypomethylating effect of decitabine treatment reactivated the lytic cycle of the latent EBV in the cancer cells. Because lysis is lethal to cells, the epigenetic reactivation of lysis within gastric cancer associated with EBV offers a promising potential treatment for the specific subset of EBVaGC.
"Now we know that we can use the epigenome of Epstein-Barr Virus against the gastric cancer that it affects—that's an exciting potential cancer therapy we have as a result of investigating the interplay between epigenetic patterns and disease lifecycle," explained Tempera.
More information: Sarah Preston-Alp et al, Decitabine disrupts EBV genomic epiallele DNA methylation patterns around CTCF binding sites to increase chromatin accessibility and lytic transcription in gastric cancer, mBio (2023). DOI: 10.1128/mbio.00396-23