This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Using large-scale molecular simulations to improve the way we prepare for future COVID-19 variants
Scientists are making significant leaps towards understanding and predicting the future threats posed by the evolution of the COVID-19 virus, thanks to powerful simulation and machine learning techniques.
The study, published in the Biophysical Journal, was led by a team of researchers from the Science and Technology Facilities Council's (STFC) Hartree Center and IBM, at the Hartree National Center for Digital Innovation (HNCDI).
It sheds valuable early insights into how viral proteins behave at the molecular level, and the physical factors responsible for viral evolution.
All viruses, including the SARS-CoV-2, which was responsible for the COVID-19 pandemic, naturally undergo genetic mutations throughout their life cycle.
While most of these mutations have minimal impact on the virus, some can alter its characteristics substantially, affecting its transmission and disease severity.
Moreover, mutations can also impact how well our vaccines and tests work, and if they provide an advantage to the virus, they can give rise to dominant variants with heightened global health risks.
Anticipating viral mutations
Significant progress has already been achieved in establishing systems to detect early signs of 'variants-of-concern' and to swiftly evaluate their impact on public health.
However, equally vital, is gaining a deeper understanding of these mutations at the molecular level and how they provide the virus with an evolutionary advantage.
These efforts, combined with other tools, hold the strong potential to anticipate and prepare for future, yet-unseen mutations and variants.
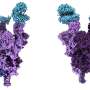
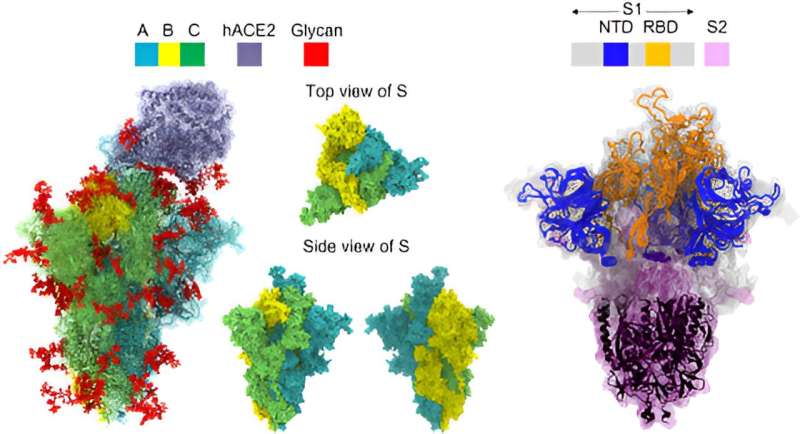
Using the powerful supercomputers at the HNCDI, the team used large-scale molecular simulations to investigate the configuration of the SARS-CoV-2 viral spike protein (responsible for causing viral infections), and how it interacts with its target on human cells.
The study highlighted how genetic changes in the omicron variants lead to structural differences at the molecular level in the spike protein, when compared to the original Wuhan strain, and even within two different omicron subvariants.
They also examined glycans, molecules connected to our cells which can influence how the virus interacts with our cells. Their findings revealed that mutations can trigger glycans to play a role in the binding process, and can make the virus bind more effectively to human cells, potentially explaining its rapid spread.
Driving a digital future
Located at STFC's Daresbury Laboratory, at Sci-Tech Daresbury in the Liverpool City Region, the Hartree Center is home to some of the UK's most advanced supercomputing experts and technologies.
David Bray, Team Leader for Chemistry and Materials at the STFC Hartree Center said, "COVID-19 has proven to be one of most infectious viruses of modern times. Determining how the virus binds to human receptors is key to understanding its high infection rate and takes us a step forward to being able to combat it and future viruses. Working alongside IBM researchers has enabled us to develop new insight into the evolutionary drivers of this key viral structure."
Ya-Wen Hsiao a Computational Scientist from the Hartree Center said, "In developing our knowledge of how the molecular mechanism works, we can contribute to the understanding of COVID-19 virus infection and help the design of vaccines or drugs. I look forward to continued research collaborations to gain a complete picture of the SARS-CoV-2 virus and its interaction with human cells."
Anticipating future variants
Professor Jason Crain from IBM Research Europe and University of Oxford, said, "The COVID-19 threat has demonstrated very clearly and quickly the consequences of viral evolution. However, the molecular mechanisms that bring advantages to the virus and select for evolutionary pathways have been under-explored. This collaboration between IBM and STFC is a promising step in that direction which may enable us to better anticipate and prepare for future variants."
These findings build upon earlier collaborative work by this team with the University of Oxford and the Diamond Light Source to use advanced artificial intelligence techniques to discover new molecular designs to inhibit the function of COVID-19 viral proteins.
More information: Ya-Wen Hsiao et al, Structure adaptation in Omicron SARS-CoV-2/hACE2: Biophysical origins of evolutionary driving forces, Biophysical Journal (2023). DOI: 10.1016/j.bpj.2023.09.003