This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New model enhances precision in seizure localization for epilepsy patients
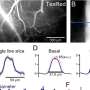
A research team led by Prof. Zhan Yang from the Shenzhen Institute of Advanced Technology (SIAT) of the Chinese Academy of Sciences, has recently introduced a novel unsupervised dual-stream model based on adaptive graph convolution to predict seizure onset zones in epilepsy patients. The study is published in Neuroimage.
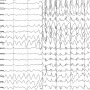
Epilepsy is a chronic disorder of the brain. One form of this disorder is refractory epilepsy characterized by persistent and drug-resistant seizures, which often requires brain surgery for the removal of the epileptic onset zone. In current clinical practice, surgical decisions rely heavily on stereoelectroencephalography (SEEG) monitoring during seizure episodes, which neglects valuable data from the rest and sleep states. The cortical–cortical evoked potential (CCEP) technique, effective in measuring synaptic connections between brain regions, is also underused in seizure localization.
To address the clinical challenges, the researchers conducted personalized analyses of SEEG (sleep, rest, and seizure states) and CCEP datasets for each patient and constructed a data-driven dual-stream deep network model. This approach sidesteps the complexities of data labeling with traditional machine learning formulations, providing patient-specific, personalized results for epilepsy focus localization.
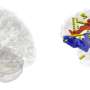
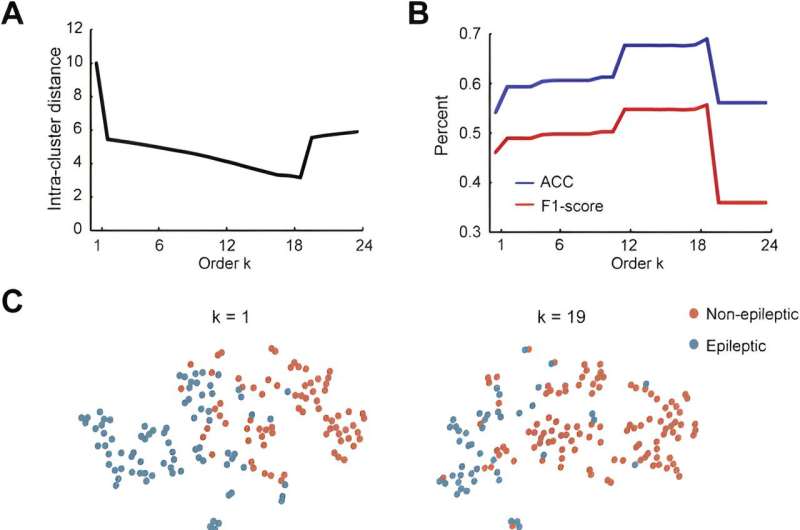
The proposed model abstracted the brain into a graph attribute network, treating each brain region as a node and the effective connections between areas as edges. The intracranial EEG recordings were used to build features for nodes, while CCEP was employed to establish topological connection relationships. Through time-frequency feature extraction and an unsupervised autoencoder, the dimensions of the features were reduced. The adaptive graph convolution network then aggregated features of adjacent nodes, ultimately classifying the location of epileptic brain regions.
In addition, the researchers delved into group-level network dynamics, and examined network characteristics between classified epileptic and non-epileptic brain areas. The results showed that brain network disparities among different epilepsy types (frontal lobe epilepsy, temporal lobe epilepsy, parietal lobe epilepsy), aligning with clinical observations.
"This study autonomously determines seizure onset zones based on individual patients' SEEG data, providing a more precise and personalized treatment approach for refractory epilepsy patients," said Prof. Zhan.
More information: Yonglin Dou et al, Identification of epileptic networks with graph convolutional network incorporating oscillatory activities and evoked synaptic responses, NeuroImage (2023). DOI: 10.1016/j.neuroimage.2023.120439