This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New method improves detection of potential therapeutic tumor targets in human biopsies
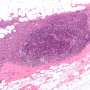
![The kinase inhibitor pulldown assay. A structure of 9 kinase inhibitors used for KiP. For afatinib, axitinib, AZD4547, and FRAX597, a C3 Linker (yellow line) is added for conjugation. The amine group for conjugation is marked with an asterisk (*). B Workflow for the kinase inhibitor pulldown assay. Native protein lysates are incubated with kinase inhibitor-conjugated beads for 1 h, and non-specific bound proteins are washed with high salt containing buffers. Inhibitor-bound kinases are digested with trypsin overnight, and digested peptides are cleaned with a detergent-removal kit and analyzed by mass spectrometry using a hybrid DDA/PRM mode. C Clustering of protein kinases enriched by individual inhibitors. Single inhibitor bead pulldown was carried out in triplicates for the 6 reference cell line mixture (6REF). Hierarchical clustering analysis of kinases in these experiments clearly shows that each kinase inhibitor pulls down distinct pools of kinases. Kinase classification by illuminating the Druggable Genome (IDG) Target Development Level (IDG-TDL) is indicated with different colors [26]. Green: Tbio, orange: Tchem, blue: Tclin, black: Tdark. FunCats are an in-house annotation of Functional Categories for different kinase targets including lipids (KI-L), metabolite (small molecule) (KI-M), proteins (KI-P) and unknown (KI-X). FunCats mapping can be found in the Additional file 2. D The Kinome tree with identified kinases highlighted. Colors represent IDG TDL classifications, and the size of the circle represents number of inhibitors that can pull down that kinase. Image rendered with KinomeRender [25]. Green: Tbio, orange: Tchem, blue: Tclin, black: Tdark. Original kinome tree illustration reproduced courtesy of Cell Signaling Technology, Inc. Credit: Clinical Proteomics (2024). DOI: 10.1186/s12014-023-09448-3 Method improves detection of potential therapeutic tumor targets in human biopsies](https://scx1.b-cdn.net/csz/news/800a/2024/method-improves-detect.jpg)
Many cancers, including some types of breast cancer, are driven by alterations in the activity of cellular enzymes called kinases. Therapies that directly inhibit these cancer-promoting activities have proven to be effective for patients in which individual driving kinases can be diagnosed.
One major challenge to this therapeutic approach is to accurately quantify tumor kinases in human biopsy samples. Many kinases are not abundantly present and are therefore more difficult to measure accurately. Although currently there are methods to quantify small amounts of kinases, measuring multiple kinases concurrently is cumbersome and impractical in a clinical setting where rapid data return is critical. It is crucial to develop methodologies to enrich kinases present in clinical samples, an important step toward effective personalized medicine.
In a study published in Clinical Proteomics, researchers at Baylor College of Medicine and collaborating institutions report the development of a kinase inhibitor pulldown assay (KiP) that can optimally enrich and quantify the small amounts of kinases present in biopsy samples in combination with mass-spectrometry techniques.
The researchers established the coverage and quantitative fidelity of the assay for kinases in a single-shot approach, optimized a 100-kinase targeted panel and determined the effectiveness of KiP in subtyping breast cancer patient-derived animal models and two breast cancer patient sample cohorts.
"Our study represents a convergence of advanced technologies, redefining basic medical research and paving the way for future clinical applications," said first author Dr. Alexander Saltzman, senior bioinformatics analyst at the Mass Spectrometry Proteomics Core at Baylor.
"This paper emphasizes that new methods in protein mass spectrometry hold great promise for better definition of the individual druggable landscape present in each cancer and should be more widely used for research, and ultimately, clinical care," said co-corresponding author Dr. Matthew Ellis, faculty at Baylor's Lester and Sue Smith Breast Center.
"This methodology's approach to identifying key kinases in cancer may even extend beyond these enzymes and into other low-abundance and biologically relevant targets," said co-corresponding author Dr. Beom-Jun Kim, currently an associate director at AstraZeneca and an assistant professor at Baylor at the time of research.
Doug W. Chan, Matthew V. Holt, Junkai Wang, Eric J. Jaehnig, Meenakshi Anurag, Purba Singh and Anna Malovannaya also contributed to this work. The authors are affiliated with Baylor College of Medicine, the Lester and Sue Smith Breast Center and/or the Dan L Duncan Comprehensive Cancer Center.
More information: Alexander B. Saltzman et al, Kinase inhibitor pulldown assay (KiP) for clinical proteomics, Clinical Proteomics (2024). DOI: 10.1186/s12014-023-09448-3