This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Network analysis provides an integrated view of multiple sclerosis

An international research team has developed a computational biology tool, based on multi-level network analysis, to achieve an integrated vision of multiple sclerosis. This tool could be used to study other complex diseases such as types of dementia.
Multiple sclerosis is an autoimmune disease of unknown cause that occurs when the immune system attacks the brain and spinal cord. It is a complex disease that is not always easy to diagnose and covers a wide range of biological scales, ranging from genes and proteins to cells and tissues, passing through the entire organism.
Symptoms of multiple sclerosis vary among patients, but the most common range from vision problems, asthenia, difficulty walking and keeping balance, to numbness or weakness in the arms and legs. All of them can appear and disappear or last over time.
The study was led by the Department of Medicine and Life Sciences (MELIS) at Pompeu Fabra University, in collaboration with Hospital del Mar, Hospital Clínic, Charité—Medical University of Berlin, and the universities of Oslo and Genoa. The study is published in PLoS Computational Biology.
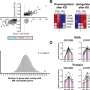
The researchers conducted a multi-level network analysis of multiomic data (genomic, phosphoproteomic and cytomic), brain and retinal images and clinical data of 328 patients with multiple sclerosis and 90 healthy subjects. It is one of the first studies to date that simultaneously analyzes data from very different scales, covering everything from genes to the whole organism. Thus, the new tool allows researchers to understand the complexity of chronic diseases.
"In this study we have analyzed five levels at once: genes, proteins, cells, parts of the brain and behavior. The proximity of the elements of each level in each person has determined the connection between the elements within each level and between levels and, through Boolean dynamics, considering each element as being active or inactive, and the introduction of disturbances in the system, we have made the elements of the network oscillate," says Jordi Garcia-Ojalvo, professor of Systems Biology and director of the Dynamical Systems Biology Laboratory at the UPF Department of Medicine and Life Sciences.
"Thus, we have managed to identify which elements of the different levels are related at the biological level."
"In complex diseases, as in society, many things happen at once, and they do so on multiple scales and over time. So, for human beings, researchers and physicians, it is hard to visualize it if it is not by using these types of tools that allow us to discern and identify the related elements," says Pablo Villoslada, an associate professor at the UPF Department of Medicine and Life Sciences, director of the Neurosciences program of the Hospital del Mar Research Institute and head of the Neurology Service at Hospital del Mar, who co-led the study together with Garcia-Ojalvo.
Thanks to the enormous capacity of networks to simplify complex data, they have managed to reveal the correlation between the protein MK03, previously associated with multiple sclerosis, with the total count of T cells, immune system cells that help fight infections, the thickness of the layer of retinal nerve fibers and the timed gait test, which measures the time it takes a patient to walk 7.5 meters as quickly as possible.
Although the size of the study has not allowed for validating the use of this correlation as a biomarker to diagnose and possibly treat multiple sclerosis, it has allowed an integrated view of this complex system and revealed the relationship between four biological scales: proteins, cells, tissues and behavior.
"In complex diseases it is very difficult to have genetic biomarkers. They are often determined by multiple genes and there is a lot of 'background noise.' And here we are studying sets of genes, proteins, and phenotypes, and if they are related to each other, we have an indication of the existence of the disease," Garcia-Ojalvo adds.
"With multiple sclerosis we have to build a puzzle whose aspect we can more or less intuit. We are not totally in the dark, which is why we use systems biology, which informs us of the relevant relationships between the elements so that the puzzle is coherent, fits and we learn. And once we know how the disease works, we can find out how to deal with it," Villoslada concludes.
This tool based on the relationship between basic biology and applied medicine could be applied to the study of other complex diseases such as Alzheimer's and other types of dementia.
More information: Multiscale networks in multiple sclerosis, PLoS Computational Biology (2024). journals.plos.org/ploscompbiol … journal.pcbi.1010980