This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
PROX1/α-SMA correlated with colorectal cancer progression, poor outcomes and therapeutic resistance
A new research paper titled "PROX1 interaction with α-SMA-rich cancer-associated fibroblasts facilitates colorectal cancer progression and correlates with poor clinical outcomes and therapeutic resistance" has been published in Aging.
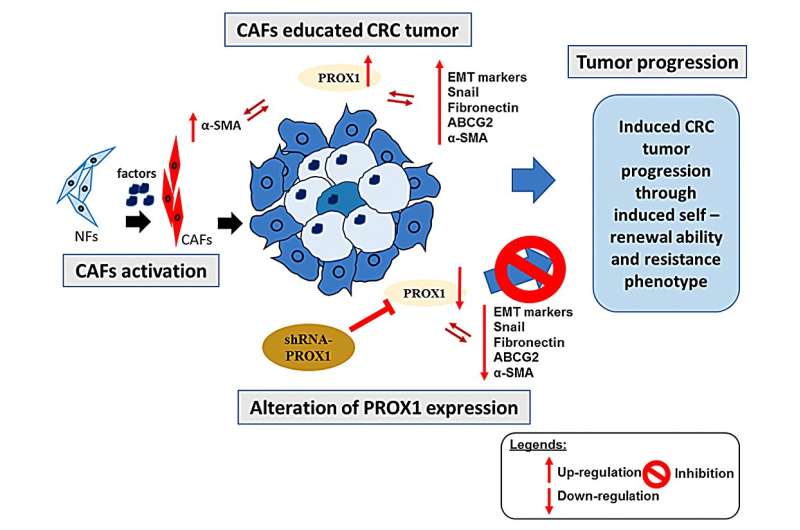
The tumor microenvironment (TME) plays a vital role in tumor progression through intricate molecular interactions. Cancer-associated fibroblasts (CAFs), notably those expressing alpha-smooth muscle actin (α-SMA) or myofibroblasts, are instrumental in this context and correlate with unfavorable outcomes in colorectal cancer (CRC).
While several transcription factors influence TME, the exact regulator causing CAF dysregulation in CRC remains elusive. Prospero Homeobox 1 (PROX1) stands out, as its inhibition reduces α-SMA-rich CAF activity. However, the therapeutic role of PROX1 is debated due to inconsistent study findings.
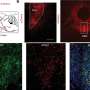
In this new study, researchers from Taipei's National Defense Medical Center, Taipei Medical University, Taipei Medical University Shuang-Ho Hospital, and National Taitung University used the ULCAN portal and noted an elevated PROX1 level in advanced colon adenocarcinoma, linking to a poor prognosis. Their assays determined the impact of PROX1 overexpression on CRC cell properties, while co-culture experiments spotlighted the PROX1-CAF relationship. Molecular expressions were validated by qRT-PCR and Western blots, with in vivo studies further solidifying the observations.
"Our study emphasized the connection between PROX1 and α-SMA in CAFs," the researchers say.
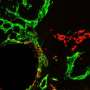
Elevated PROX1 in CRC samples correlated with increased α-SMA in tumors. PROX1 modulation influenced the behavior of specific CRC cells, with its overexpression fostering invasiveness. Kaplan-Meier evaluations demonstrated a link between PROX1 or α-SMA and survival outcomes. Consequently, PROX1, alone or with α-SMA, emerges as a CRC prognostic marker. Co-culture and animal experiments further highlighted this relationship.
PROX1 appears crucial in modulating CRC behavior and therapeutic resistance within the TME by influencing CAFs, signifying the combined PROX1/α-SMA gene as a potential CRC prognostic marker. The concept of developing inhibitors targeting this gene set emerges as a prospective therapeutic strategy. However, this study is bound by limitations, including potential challenges in clinical translation, a focused exploration on PROX1/α-SMA potentially overlooking other significant molecular contributors, and the preliminary nature of the inhibitor development proposition.
"As we advance in this field, the development and clinical validation of small-molecule inhibitors targeting PROX1/α-SMA become imperative, paving the way to refine and optimize CRC therapeutic interventions," the researchers conclude.
More information: Shiue-Wei Lai et al, PROX1 interaction with α-SMA-rich cancer-associated fibroblasts facilitates colorectal cancer progression and correlates with poor clinical outcomes and therapeutic resistance, Aging (2024). DOI: 10.18632/aging.205447