This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers may have found key to deprogram cells that lead to transplant rejection
Houston Methodist researchers identified a troublesome subset of T-cells in transplant recipients that may be a more effective therapeutic target for preventing transplant rejection in patients.
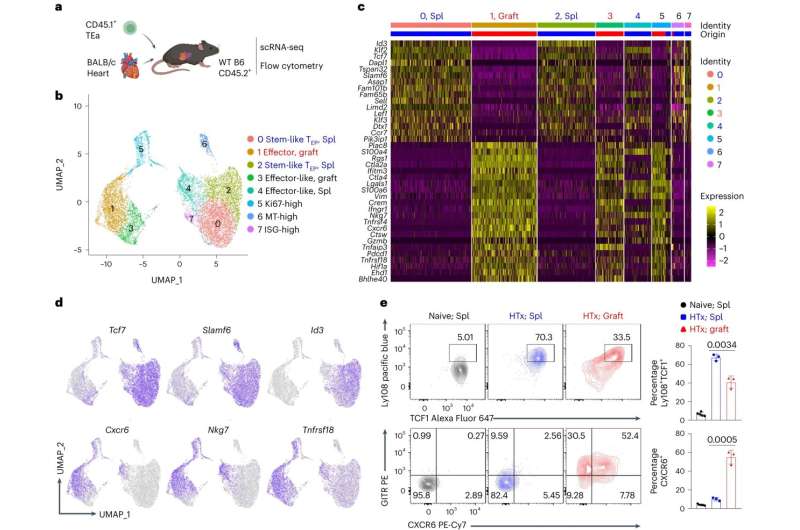
Each day, 17 people die waiting for organ transplants, but getting a new organ doesn't guarantee survival. Despite immunosuppressive medications, rejection of transplanted organs happens in up to 50% of patients, depending on the type of organ transplanted and the time since the transplantation.
Wenhao Chen, Ph.D., associate professor of transplant immunology with the Houston Methodist Research Institute, and his team used single-cell RNA sequencing to analyze the CD4+ T-cell response in transplantation scenarios and identified a subset that he referred to as "troublemakers." They also discovered the mechanism behind what directs that T-cell response in animal transplant models.
This troublesome subset of CD4+ T-cells they discovered acts like stem cells and continuously generates functionally mature effector T-cells that attack transplanted organs. They also found that the transcription factor IRF4 is required for T-cells in that subset to become those organ-attacking effector T-cells. Thus, Chen says IRF4 is what needs to be targeted to solve the problem of transplant rejection or to develop an autoimmunity cure.
"T-cells play a central role in fighting infections and cancer, but they are also the key players in mediating autoimmune diseases and transplant rejection," Chen said. "Our study demonstrated that IRF4 is a master regulator of T-cell function; a discovery that will allow the development of innovative therapies for patients with chronic infections, cancers, autoimmune diseases, and transplanted organs."
Eliminating undesired CD4+ T-cell responses that could potentially lead to the loss of transplanted organs could eventually apply to all transplant patients, he said of this discovery.
"How to therapeutically inhibit IRF4 is the Nobel-prize winning question," Chen said in reference to a past research study on this challenge. "If we can find a way to inhibit IRF4 as desired in activated T-cells, then I think most autoimmune diseases and transplant rejection will be solved."
Their findings are described in an article titled "CD4+ T cell immunity is dependent on an intrinsic stem-like program" in a recent issue of Nature Immunology.
"This revelation about the true troublemaker within the CD4+ T-cell population is just the tip of the iceberg," Chen said. "I sincerely hope our findings garner widespread attention, motivating both researchers and patients to recognize the significance of targeting these troublemakers."
More information: Dawei Zou et al, CD4+ T cell immunity is dependent on an intrinsic stem-like program, Nature Immunology (2024). DOI: 10.1038/s41590-023-01682-z