This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Using machine learning to track the evolution of COVID-19
Scientists have developed a machine-learning approach to track the evolution of SARS‑CoV‑2, the virus that causes COVID-19, and potentially other viruses, according to a study published in the Proceedings of the National Academy of Sciences.
Since the onset of the COVID-19 pandemic, 1,185,413 people in the United States have died from the virus, according to data collected by the Centers for Disease Control and Prevention.
RNA viruses, such as SARS‑CoV‑2, mutate rapidly once inside a host. Most RNA viruses including HIV-1 or influenza acquire a high number of mutations to the point where, in many cases, no two copies of the virus inside one person have the exact same genome.
These mutated strains can then jump to the general population, pushing forward the evolution of these viruses at a global level. While SARS‑CoV‑2 has been reported to mutate at lower rates compared to similar viruses, it has shown a high capacity to evolve with new variants appearing suddenly instead of progressively.
This observation challenges the previous idea of low mutation capacity of SARS-CoV-2, said Ramon Lorenzo-Redondo, Ph.D., assistant professor of Medicine in the Division of Infectious Diseases and bioinformatics director of the Center for Pathogen Genomics and Microbial Evolution (CPGME), who was a co-author of the study.
The origins of these highly mutated variants such as omicron, which acquired a very high number of mutations rapidly, is still poorly understood, he said.
In the study, Lorenzo-Redondo and his collaborators applied a novel next-generation sequencing method to sequence the genome of SARS-CoV-2 from thirty individual nasal swab samples obtained within a 19-day window.
With this new method, developed by senior study author Esteban Domingo, Ph.D., and study co-author Celia Perales, Ph.D., investigators at the Spanish National Research Council, the team was able to capture a wide representation of every mutant of the virus present inside each patient. This way, they could study if minority mutations generated inside an infected patient could be the origin of mutations that later get transferred to the general population.
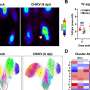
Then, utilizing a machine-learning model first developed by Lorenzo-Redondo and Soledad Delgado, Ph.D., associate professor at the Polytechnical University of Madrid, the investigators visualized genetic data from the samples into maps which showed the many variations of the virus inside a single host and charted their predicted survival and proliferation in relation to the other variants.
The technique may allow scientists to track how viruses like SARS-CoV-2 evolve over time inside a single person and predict dangerous mutations, Lorenzo-Redondo said.
"With this technique, we can go deeper. We can analyze evolution and analyze how the virus is adapting to a person and how it evolves to counter the immune system," Lorenzo Redondo said. "Some of these adapted viruses then might become important at a population level."
By sequencing SARS-CoV-2 inside an individual, investigators observed how the virus "tested" a mutation in its spike protein in some individuals, which has been reported to alter viral entry. This specific spike protein mutant variant was a small subset inside of some of these hosts, but subsequently, it quickly overtook other variants due to its superior infectivity and became the dominant strain globally during the first months of the pandemic.
The findings may explain how new variants materialize and jump from person to person and become dominant, strengthening the entire viral population, Lorenzo Redondo said.
"This is very interesting because it seems to suggest that all these big jumps that we see, for example in the omicron wave of COVID-19, might be happening at the intra-host level in multiple patients at the same time and then get transferred to the general population," he said.
Moving forward, Lorenzo Redondo and his collaborators will aim to use this combination of novel molecular biology techniques and machine learning approaches to map intra-host evolution in SARS-CoV-2 and other viruses, he said. His group also hopes to use the approach to predict how a virus may evolve in the future and potentially stop dangerous strains from taking hold.
"The next step is: can we use machine learning methods to predict possible future mutations by knowing what the virus has already explored and what type of advantage it gave it inside the host?" Lorenzo Redondo said.
More information: Soledad Delgado et al, Incipient functional SARS-CoV-2 diversification identified through neural network haplotype maps, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2317851121