This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New method rapidly reveals how protein modifications power T cells
Imagine riding a bike. Now imagine riding a bike with an enormous beach ball stuck on your handlebars. That "modification" might change your experience quite a bit. In our cells, molecules called phosphate groups are constantly attaching to—or detaching from—proteins. These "post-translational modifications" alter how our proteins function.
"Small kinds of chemical 'ornaments' are added or removed from proteins in a very rapid fashion, and these modifications change how a protein interacts with other biomolecules," says La Jolla Institute for Immunology (LJI) and Global Autoimmune Institute Assistant Professor Samuel Myers, Ph.D.
Myers leads LJI's Laboratory for Immunochemical Circuits, and his team has pioneered the use of mass spectrometry, advanced genome engineering tools, and phenotypic screens to study protein modifications with important roles in immune cells. Myers and his colleagues recently developed a new method, published in Nature Methods, to quickly assess the "phosphorylation sites" where phosphate groups attach to proteins and modulate their function.
This method is a major leap forward for scientists investigating how proteins do their jobs in the immune system. "This method basically creates an entire new kind of world of experiments that people can do," adds Myers.
For the new study, the researchers used the method to shed light on proteins that form complex "signaling pathways" in disease-fighting T cells.
"There's so much we don't know about these signaling pathways," says LJI Research Technician Patrick Kennedy, who served as first author of the new study. "This new method opens the door for researchers to learn more."
How the new method works
To make a protein, a cell "transcribes" DNA into a useful molecule called mRNA. Cells then "translate" the mRNA code to assemble proteins. The proteins then get modified to achieve their final active form. For the new study, Myers and his team focused on the modifications that make up the signaling pathways that trigger T cell activation.
T cell activation is when T cells ready themselves to fight a pathogen. "T cell activation is at the crux of many autoimmune disorders, and problems with T cell activation are the reason why cancer doesn't get taken care of properly by the immune system," says Myers. "So we need a higher resolution understanding of signaling and T cell activation pathways."
T cells coordinate huge batches of new post-translational modifications during the activation process. Myers and his colleagues wanted to know exactly where phosphate groups attach themselves to these proteins. How might these modifications influence function? Would they increase or decrease protein function—and would there be consequences for T cell activation?
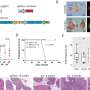
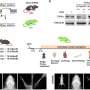
To find out, the scientists harnessed a gene editing tool called Cas9-mediated base editors, which they used to make very small, targeted tweaks to a T cell's DNA code. This gave the scientists a library of mutated proteins that can no longer be modified in the same way. They then prompted the mutant T cells to assemble based on their altered function.
Because of the mutated DNA—and eventually, RNA—these new proteins lacked their usual phosphorylation sites. Myers and his team then tested whether the new proteins had a decrease or increase in function—or if the changes led to any functional differences at all.
The new method led them to answers—at lightning speed. "People have typically looked at 10 or 20 phosphorylation sites at a time," says Myers. "But because we're a mass spectrometry lab, we can use this technique to look at all phosphorylation—and we can now do this functional testing for 10,000 or more phosphorylation sites at a time."
The scientists quickly analyzed around 11,000 phosphorylation sites. Then they worked with the LJI Next Generation Sequencing Core to profile the edited T cells. Their analysis revealed how phosphate groups direct specific gene expression responses as the cell prepares to fight disease.
"Depending on what mutation we make, we might be able to actually tune the levels of different subsets of genes within the T cell activation pathway," says Myers.
Going forward, Myers hopes researchers can take genetic "tuning" a step further and figure out how to activate specific immune cells to fight disease. For example, scientists might wish to activate only certain T cell subsets to stop cancer development. "Could we activate CD8+ 'killer' T cells in the context of treating cancer without activating the regulatory T cells which normally dampen the immune system?" says Myers.
Phosphate groups aren't the only molecules that alter protein function, adds Myers. As he explains, the new method can apply to studying many types of post-translational modifications, including a modification Myers called his "first love," O-GlcNAc.
Additional authors of the study, "Post-translational modification-centric base editor screens to assess phosphorylation site functionality in high-throughput," included Amin Alborzian Deh Sheikh, Matthew Balakar, Alexander C. Jones, Meagan E. Olive, Mudra Hegde, Maria I. Matias, Natan Pirete, Rajan Burt, Jonathan Levy, Tamia Little, Patrick G. Hogan, David R. Liu, John G. Doench, Alexandra C. Newton, Rachel A. Gottschalk, Carl de Boer, Suzie Alarcón, and Gregory Newby.
More information: Patrick H. Kennedy et al, Post-translational modification-centric base editor screens to assess phosphorylation site functionality in high throughput, Nature Methods (2024). DOI: 10.1038/s41592-024-02256-z