This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
New insights into biomarkers and probes for prostate cancer
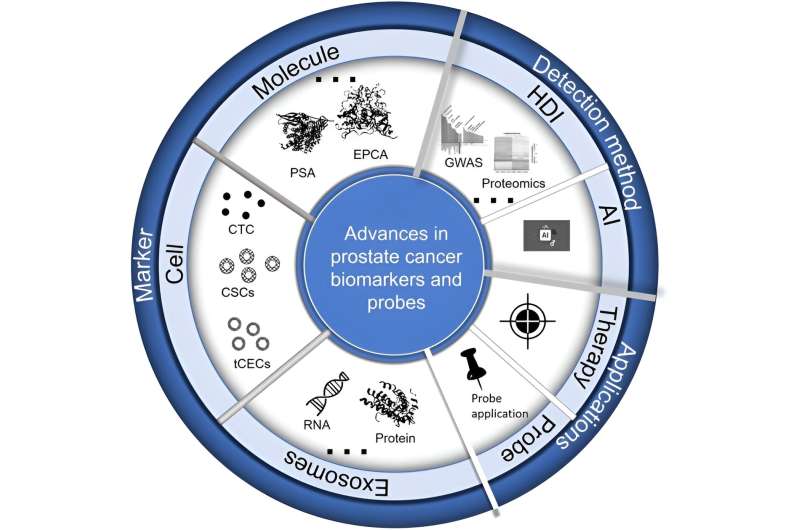
In a recent comprehensive review published in Cyborg Bionic Systems, researchers detail significant advances in the identification and application of biomarkers for prostate cancer (PCa). This critical insight is pivotal as prostate cancer remains one of the most common malignancies among men globally, emphasizing the urgent need for effective diagnostic and therapeutic strategies. The team was led by Keyi Li from the General Hospital of Northern Theater Command in Shenyang, along with international collaborators,
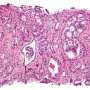
Prostate cancer is characterized by a multitude of molecular aberrations that complicate its early detection and treatment. The review discusses various biomarkers across molecular, cellular, and exosomal categories, showcasing their potential to revolutionize how prostate cancer is diagnosed and treated.
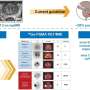
One of the major highlights of the review is the discussion on gene fusions like TMPRSS2-ERG, which are prevalent in prostate cancer and serve as vital diagnostic indices. The presence of noncoding RNAs such as SNHG12 and proteins like PSA and PSMA also hold substantial promise in not only detecting the disease but also in understanding its progression and guiding therapy.
In terms of technological integration, the review illuminates the growing role of multi-omics data and artificial intelligence in prostate cancer research. These technologies contribute significantly to the biomarker discovery process, enhancing the ability to tailor personalized medicine approaches for patients. For instance, AI and genomics are being used to better predict disease outcomes and respond to treatment regimens, highlighting a shift towards more precise and personalized cancer care.
Moreover, the development of specific probes for biomarker detection is another area of rapid advancement. The review details innovations in creating fluorescent, electrochemical, and radionuclide probes that enhance the sensitivity and specificity of current diagnostic techniques.
The review also delves into the implications of these discoveries for the future of prostate cancer treatment. With these biomarkers and technological advancements, there is a clear path toward more targeted therapies, which could lead to better patient outcomes and less invasive treatment options.
This extensive analysis not only serves as a valuable resource for health care professionals and researchers but also provides hope for patients by advancing our understanding of prostate cancer at the molecular level. The integration of these cutting-edge technologies and discoveries promises a future where prostate cancer can be diagnosed more accurately, treated more effectively, and managed with a precision that was previously unattainable.
More information: Keyi Li et al, Advances in Prostate Cancer Biomarkers and Probes, Cyborg and Bionic Systems (2024). DOI: 10.34133/cbsystems.0129