Team creates new view of body's infection response
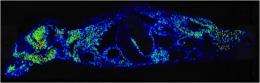
A new 3-D view of the body's response to infection – and the ability to identify proteins involved in the response – could point to novel biomarkers and therapeutic agents for infectious diseases.
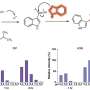
Vanderbilt University scientists in multiple disciplines combined magnetic resonance imaging (MRI) and imaging mass spectrometry to visualize the inflammatory response to a bacterial infection in mice. The techniques, described in Cell Host & Microbe and featured on the journal cover, offer opportunities for discovering proteins not previously implicated in the inflammatory response.
Access to unique resources at Vanderbilt made the unprecedented 3-D infection imaging possible, said Eric Skaar, Ph.D., Ernest Goodpasture Chair in Pathology and one of the senior co-authors of the paper.
"The studies in this paper couldn't have happened at any other university, because the resources simply don't exist at most schools," Skaar said.
The resources include animal imaging technologies available through the Vanderbilt University Institute of Imaging Science (VUIIS), directed by John Gore, Ph.D., and imaging mass spectrometry technologies available through the Mass Spectrometry Research Center (MSRC), directed by Richard Caprioli, Ph.D. Gore and Caprioli are also senior co-authors of the paper.
"The fact that my research group, which studies infectious diseases, has access to these powerful imaging and mass spectrometry technologies is a real strength at Vanderbilt and has allowed us to develop these new tools that will enable high impact discovery," Skaar said.
Skaar and his team were interested in imaging infection in three dimensions – in the whole animal – while also being able to identify the proteins that are produced at sites of infection. MRI provides detailed anatomical images of tissue damage. Imaging mass spectrometry is a unique technology that directly measures proteins, lipids and other metabolites and maps their distribution in a biopsy or other tissue sample.
Ahmed Attia, Ph.D., a former member of Skaar's group now on the faculty at Cairo University, Egypt, infected mice with Staphylococcus aureus, a major cause of human disease. He then delivered the infected animals to Daniel Colvin, Ph.D., in the VUIIS, who imaged them with MRI. Kaitlin Schroeder and Erin Seeley, Ph.D., in the MSRC then conducted imaging mass spectrometry studies. Putting together the two technologies and multiple data sets accurately required the expertise of Kevin Wilson, MESc, in the VUIIS, who developed algorithms to show consolidated 3-D views of the inflammatory response.
"This is another example of the multi-modality approach we have been pursuing in general within the Imaging Institute," Gore said.
The technologies allow the investigators to see a single image of an infected animal, look at how proteins of the immune system are responding, and identify where the infected tissue is located, Skaar said.
"Part of the strength of this work is not where the research is now, but where it allows us to go from here."
His team plans to identify "proteins that are important at the interface between the host and the pathogen – the battleground between the immune system and the bacteria," Skaar said. The researchers will study the proteins they identify to discover new biomarkers for infection, which could improve diagnostic tools, or new targets for therapeutic intervention.
The technologies available through the MSRC and the VUIIS will be useful for any investigator interested in imaging the inflammatory response, which has roles in infectious diseases, cancer and autoimmune diseases, Skaar said. And although the technology is not non-invasive (imaging mass spectrometry requires tissue sections), it could be applied to tissues removed from patients, such as tumors.
"Imaging mass spectrometry is extremely valuable for the discovery process because it does not require a target-specific reagent such as an antibody – that is, you do not have to know in advance what you're looking for in order to correlate molecular changes with disease outcome," Caprioli said. "An area of intense interest is the application of this technology to molecular pathology."
More information: www.cell.com/cell-host-microbe … 1931-3128(12)00165-5