Ovarian cancer study provides painstaking look at inner workings of tumors
In what is believed to be the largest study of its kind, scientists at the Pacific Northwest National Laboratory, Johns Hopkins University and their collaborators from institutions across the nation have examined the collections of proteins in the tumors of 169 ovarian cancer patients to identify critical proteins present in their tumors.
By integrating their findings about the collection of proteins (the proteome) with information already known about the tumors' genetic data (the genome), the investigators report the potential for new insights into the progress of the most malignant form of the disease. The work is published June 29, 2016, in the advance online edition of Cell.
The researchers say their achievement illustrates the power of combining genomic and proteomic data - an approach known as proteogenomics - to yield a more complete picture of the biology of a cancer that accounts for three percent of all cancers in women and is the fifth leading cause of cancer deaths among women in the United States.
"Historically, cancer's been looked at as a disease of the genome," said Karin Rodland, a senior author of the study and chief scientist for biomedical research at PNNL, a U.S. Department of Energy laboratory. "But that genome has to express itself in functional outcomes, and that's what the proteomic data adds, because proteins do the actual work of the genome."
Daniel W. Chan, the study's other senior author, who led the team at the Johns Hopkins University School of Medicine, said, "Correlating our data with clinical outcomes is the first step toward the eventual ability to predict outcomes that reflect patient survival, with potential applications for precision medicine and new targets for pharmaceutical interventions. But just like anything in medicine, clinical validation will be a long and rigorous process."
The authors say that with the findings, researchers expect to be better able to identify the biological factors defining the 70% of ovarian cancer patients who suffer from the most malignant form of ovarian cancer, called high-grade serous carcinoma. Currently, only one in six such patients lives five or more years beyond diagnosis.
The power of collaboration
The work draws on the efforts of physicians, scientists and patients who have worked together to understand ovarian cancer. The investigators say the effort requires collaboration among physicians as well as patients willing to take part in research to benefit others with the disease or even to prevent others from ever developing cancer.
Under the leadership of the National Cancer Institute, scientists around the nation have worked together to create the Cancer Genome Atlas (TCGA), a collaborative effort to map cancer's genetic mutations. The task for ovarian cancer was completed in 2011. In the current study, the PNNL and JHU teams each studied subsets of 169 high-grade serous carcinoma tissue samples and accompanying genomic and clinical data drawn from that study.
The Johns Hopkins team initially selected 122 of the samples based on those tumors' ability to repair damaged DNA - known as homologous recombination deficiency - and characterized by changes in genes including BRCA1, BRCA2 and PTEN, mutations long linked to increased cancer risk and severity.
"We chose to examine these samples because patients with changes in these genes already are benefiting from a specific drug regimen for breast cancer, so if we could find similar changes in ovarian cancer genomes and proteomes, those patients would likely benefit from the same regimen," said Chan, a professor of pathology and oncology at JHU. Chan is one of the inventors of the OVA1 ovarian cancer detection test, which is licensed to Vermillion Inc. of Austin, Texas.
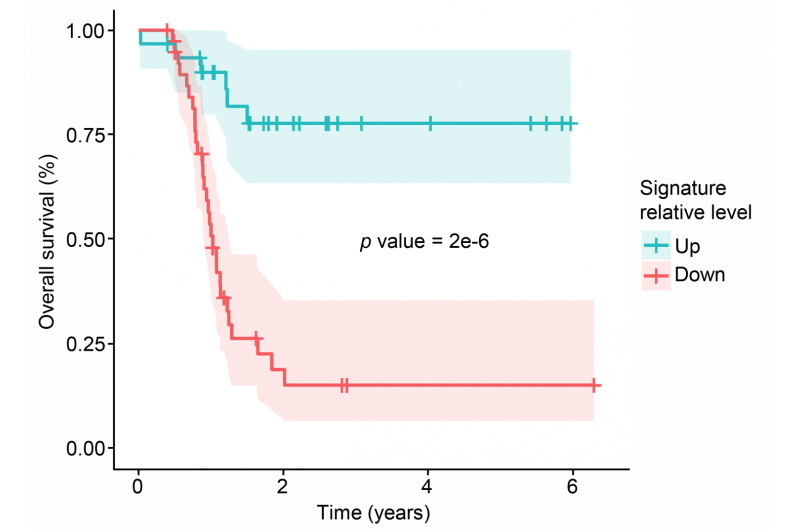
The PNNL team initially selected 84 samples based on overall patient survival times. "We examined the data for the shortest-surviving patients and the longest-surviving patients hoping to pinpoint biological factors associated with extremely short survival or better-than-average, longer survival," said Rodland.
Then, through their participation in the Clinical Proteomic Tumor Analysis Consortium (CPTAC), another program of the National Cancer Institute which funded both teams, the two groups combined their efforts.
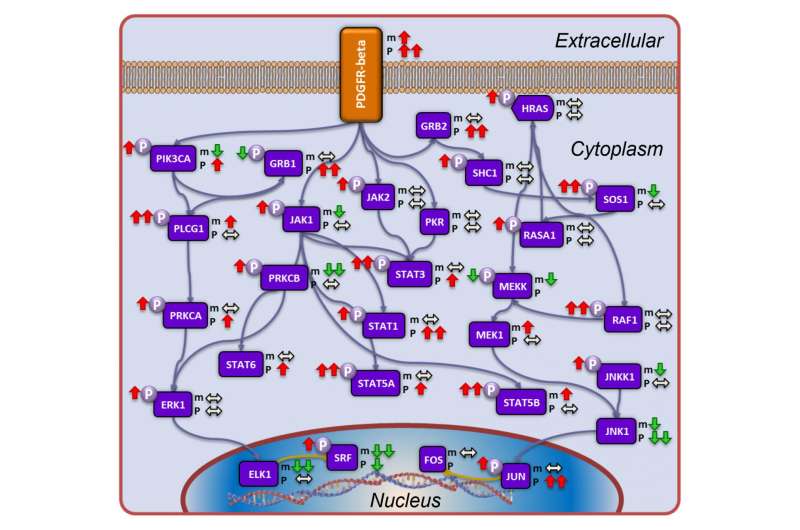
Using protein measurement and identification techniques based on mass spectrometry, the teams identified 9,600 proteins in all the tumors, and pursued study on 3,586 proteins common to all 169 tumor samples.
Beyond the genome
While many people are familiar with the role our genes play in the development of cancer, the genes are often just a starting point, for patients and researchers alike. Genes are transcribed into RNA, the genetic material that makes proteins, which are the workhorses of cells. The activity of the proteins varies dramatically, with many undergoing changes that affect their impact and interactions with other proteins.
A detailed look at the activity of proteins in cancer biology gives researchers insight into specific molecular events that would otherwise remain unknown.
A hallmark of cancer, and particularly high grade serous carcinoma, is when genetic instructions go awry. One form is the appearance of more copies of certain regions of the genome. These so-called copy number alterations can lead to changes in protein abundance. When the researchers compared known regions of copy number alterations, they found that parts of chromosomes 2, 7, 20 and 22 led to changes in abundance of more than 200 proteins. A more careful study of those 200 proteins revealed that many are involved in cell movement and immune system function, both processes implicated in cancer progression, the researchers said.
"Adding the information about the proteome on top of the genome provides an entirely new dimension of information that has enabled the discovery of new biological insights to ovarian cancer, while creating a valuable resource that the scientific community can use to generate new hypotheses about the disease, and how to treat it," said Rodland.
"High grade serous carcinoma is such a challenging disease, requiring complex clinical care to achieve long-term survival. This new knowledge gives us new directions to test in the lab and clinic," said study author Douglas A. Levine, director of gynecologic oncology at the Laura and Isaac Perlmutter Cancer Center of NYU Langone Medical Center. "This proteogenomic analysis will help us improve patient outcomes and quality of life."
In addition to large teams of scientists from PNNL and Johns Hopkins, contributors included colleagues from Stanford University School of Medicine, Vanderbilt University School of Medicine, University of California at San Diego, New York University School of Medicine, Virginia Tech, the National Cancer Institute's Office of Cancer Clinical Proteomics Research, as well as CPTAC investigators.
The proteomic analyses performed by the PNNL team were done at EMSL, the Environmental Molecular Sciences Laboratory, a DOE Office of Science user facility.
More information: Hui Zhang, Tao Liu, Zhen Zhang, Samuel H. Payne, Bai Zhang, Jason E. McDermott, Jian-Ying Zhou, Vladislav A. Petyuk, Li Chen, Debjit Ray, Shisheng Sun, Feng Yang, Lijun Chen, Jing Wang, Punit Shah, Seong Won Cha, Paul Aiyetan, Sunghee Woo, Yuan Tian, Marina A. Gritsenko, Therese R. Clauss, Caitlin Choi, Matthew E. Monroe, Stefani Thomas, Song Nie, Chaochao Wu, Ronald J. Moore, Kun-Hsing Yu, David L. Tabb, David Fenyö, Vineet Bafna, Yue Wang, Henry Rodriguez, Emily S. Boja, Tara Hiltke, Robert C. Rivers, Lori Sokoll, Heng Zhu, Ie-Ming Shih, Leslie Cope, Akhilesh Pandey, Bing Zhang, Michael P. Snyder, Douglas A. Levine, Richard D. Smith, Daniel W. Chan, Karin D. Rodland, and the CPTAC investigators, Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer, Cell, June 29, 2016, DOI: 10.1016/j.cell.2016.05.069