MicroRNA-TP53 circuit connected to chronic lymphocytic leukemia
The interplay between a major tumor-suppressing gene, a truncated chromosome and two sets of microRNAs provides a molecular basis for explaining the less aggressive form of chronic lymphocytic leukemia, an international team of researchers reports today in the Jan. 4 edition of the Journal of the American Medical Association.
"Our findings could reveal new mechanisms of resistance to chemotherapy among leukemia patients as this feedback mechanism could help us differentiate between patients with poor or good prognosis," said co-senior author George Adrian Calin, M.D., Ph.D., associate professor in The University of Texas MD Anderson Cancer Center Department of Experimental Therapeutics and co-director of the Center for RNA Interference and Noncoding RNAs.
B cell chronic lymphocytic leukemia (CLL) is the most common form of leukemia among adults, with an estimated 14,990 new cases in 2010 in the United States. It's caused by aberrant versions of infection-fighting B cell lymphocytes, a white blood cell.
Deletion of the long arm of chromosome 13, called 13q, has been associated with less aggressive, or indolent, CLL. In a series of experiments, a team led by Calin and colleagues at MD Anderson and The Ohio State University uncovered the details of that relationship.
"This finding represents the most detailed pathogenetic mechanism involving microRNAs for any human disease," Calin said. MicroRNAs, or miRNAs, are short, single-stranded bits of RNA that regulate the messenger RNA expressed by genes to tell a cell's protein-making machinery which protein to make.
Deletion of 13q unleashes two proteins known to stymie programmed cell death, or apoptosis, a frontline defense against formation and growth of malignant cells, the researchers found. However, it also leads to increased expression of the tumor suppressor gene TP53, which indirectly reduced levels of another protein associated with poor survival among CLL patients. The first effect appears to cause CLL, while the combination of effects keeps it indolent, Calin said.
The team worked with three chromosomal deletions common to CLL:
- Deletion 13q, causes the abolition or reduction of two miRNAs: miR-15a and miR-16-1. It's the most common form of deletion, occurring in about 55 percent of CLL cases.
- Deletion 11q occurs in about 18 percent of CLL cases and affects two other miRNAs: miR-34b and miR-34c.
- The short arm of chromosome 17, called 17p, is home to the tumor-suppressing gene tumor protein p53, or TP53. It is deleted in about 7 percent of CLL.
The researchers analyzed blood samples from 208 CLL patients, comparing the relative expression and activity of the miRNAs and TP53 depending on the type or types of deletions the patients had.
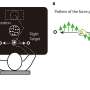
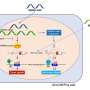
Normal Conditions, No CLL
With no chromosomal deletions, the microRNAs miR-15a and miR-16-1 inhibit expression of the tumor-suppressor TP53. However, TP53 in turn activates the two miRNAs, which then control expression of two other genes that prevent programmed cell death, or apoptosis. Apoptosis occurs at normal levels, preventing leukemia development.
TP53 also activates the miR34b and miR-34c microRNAs, which then inhibit the gene ZAP70, which is associated with shorter survival among CLL patients.
Why Indolent CLL Follows 13q Deletions
With 13q deletions, miR-15a and miR16-1 are reduced or absent, which allows for increased expression of TP53. This further activates miR-34b and miR-34a, resulting in greater inhibition of Zap70. That's the good part.
With miR-15a and 16-1 gone, expression of the two genes that suppress programmed cell death, BCL2 and MCL1, increases, which allows more aberrant cells to form and grow. While this effect causes slow-growing CLL, the counterweight of suppressed ZAP70 keeps it from getting worse, Calin said.
TP53 is, in effect, a link that joins the two different sets of microRNAs, the researchers noted.
Experiments in acute myeloid leukemia, lung and cervical cancer cell lines indicate the same mechanism may be at work in other cancer types, but more studies will be needed to validate that, Calin said.
This study clears the way for further translational research to identify microRNAs involved in resistance to CLL therapy and therefore poor outcomes for patients, as well as the identification of plasma miRNAs with predictive value for response to therapy. A team led by Calin and study co-author Michael Keating, M.D., professor in MD Anderson's Department of Leukemia, is investigating these topics.