New method may allow personalized clinical trial for cancer therapies
A new tool to observe cell behavior has revealed surprising clues about how cancer cells respond to therapy – and may offer a way to further refine personalized cancer treatments.
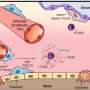
The approach, developed by investigators at Vanderbilt-Ingram Cancer Center, shows that erlotinib – a targeted therapy that acts on a growth factor receptor mutated in some lung, brain and other cancers – doesn't simply kill tumor cells as was previously assumed. The drug also causes some tumor cells to go into a non-dividing (quiescent) state or to slow down their rate of division. This variability in cell response to the drug may be involved in cancer recurrence and drug resistance, the authors suggest.
The new tool, reported Aug. 12 in Nature Methods, may offer ways to improve personalized cancer therapy by predicting tumor response and testing combinations of targeted therapies in an individual patient's tumor.
In the personalized approach to cancer treatment, a patient's tumor is analyzed for a set of mutations to which there are matching drugs that act on those mutations.
This approach has worked rather well for many cancers that carry specific mutations, said senior author Vito Quaranta, M.D., professor of Cancer Biology.
"The genetics is well understood, the clinical effect is understood and the chemistry behind the therapy is understood. But there is a missing piece," said Quaranta. "Believe it or not, what is actually not understood is how cells respond to these drugs, what is actually happening."
The prevailing view has been that targeted therapies kill all the cells harboring a particular mutation.
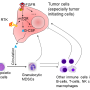
But even if the tumor is composed entirely of genetically identical cells – which is unlikely – a drug will not affect all cells the same way, Quaranta explained.
"Some of these cells may die, some may just stop dividing and sit there (called quiescence), and some may keep dividing, but more slowly."
However, no current tests can provide an accurate, detailed picture of cell behavior needed to understand tumor response to drugs.
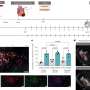
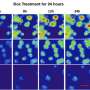
So, the investigators, led by first author Darren Tyson, Ph.D., research assistant professor of Cancer Biology, combined powerful automated, time-lapse microscopy with analytical tools and software they developed.
Using these techniques, they could capture the behavior of lung cancer cells every six to 10 minutes for up to 10 days.
As they expected, the targeted therapy erlotinib killed some cells, while others became quiescent. They observed that the drug even affected genetically identical cells (cells that arose from the same parental cell) differently.
"These cells are clearly genetically identical, as identical as they can possibly be because one cell just divided into two, but you get completely different responses: one dies and the other one doesn't," said Tyson. "This suggests that there are other things besides genetics that have to be taken into account."
What those other factors are remains unclear, but the investigators are conducting follow- up experiments to determine what might underlie this differential response.
"And presumably, it is those (quiescent) cells that ultimately result in tumor recurrence," said Tyson.
Quaranta and colleagues hope to take the technology into small clinical trials to test whether it can predict a patient's response to therapy.
"We think that we might be able to forecast what the response is going to be," Quaranta said. "We can take samples from the tumor, subject them to this assay, and since we're looking at response over time, we will have a rate of response."
This could tell oncologists how long a patient's tumor will respond to a given therapy before it recurs. Such information could also help determine which patients will require more aggressive treatment – and Quaranta believes the assay will be able to test combinations of drugs on a patient's tumor cells to find the right combination to induce a response.
"We're hoping that this assay – or some implementation of this assay – will eventually work like a personalized clinical trial," Quaranta said.