Researchers identify possible new oncogene and future therapy target
A gene that may possibly belong to an entire new family of oncogenes has been linked by researchers with the U.S. Department of Energy (DOE)'s Lawrence Berkeley National Laboratory (Berkeley Lab) to the resistance of breast cancer to a well-regarded and widely used cancer therapy.
One of the world's leading breast cancer researchers, Mina Bissell, Distinguished Scientist with Berkeley Lab's Life Sciences Division, led a study in which a protein known as FAM83A was linked to resistance to the cancer drugs known as EGFR-TKIs (Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors). Not only may this discovery explain the clinical correlation between a high expression of FAM83A and a poor prognosis for breast cancer patients, it may also provide a new target for future therapies.
"Resistance to EGFR-TKIs has limited their use for breast cancer treatment and until now the mechanisms behind this resistance have largely been a mystery," Bissell says. "We've demonstrated, both in cultured cells and in mice, that FAM83A has oncogenic properties and when overexpressed in cancer cells confers EGFR-TKI resistance and promotes the proliferation and invasion of tumors."
Bissell is the corresponding author along with Saori Furuta, also with Berkeley Lab's Life Sciences Division, of a paper describing this research in the Journal of Clinical Investigation (JCI). The paper is titled "FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice." Other co-authors are Sun-Young Lee, Roland Meier, Marc Lenburg, Paraic Kenny and Ren Xu.
Therapeutic targeting of oncogenes can be an effective way to fight some cancers as evidenced in the successful use of EGFR-TKIs to fight lung cancer. However, EGFR-TKIs have not been effective for treating breast cancers. EGFR-TKIs work by blocking EGFR from adding a phosphate molecule to downstream signaling proteins, an action called phosphorylation that is a necessary step in the development of many types of cancer.
"We hypothesized that resistance to EGFR-TKIs originated, at least in part, from molecular alterations that activated phosphorylation signaling downstream of EGFRs," Furuta says.
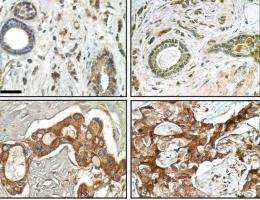
Using a unique three-dimensional cell culture assay based on phenotypic reversion that was originally developed by Bissell and her research group, the co-authors of the JCI paper screened for genes involved in EGFR-TKI resistance in both normal and cancerous human breast cell lines. They found that, while normal human breast tissue doesn't produce FAM38A, the protein is highly expressed in cancerous tissue. This was true for every breast cancer cell line they examined, and was particularly pronounced in those cell lines that have been the most resistant to treatment with EGFR-TKIs. Further studies showed that FAM83A interacts with and causes phosphorylation of critical signaling proteins downstream of EGFRs, as the researchers hypothesized. This downstream phosphorylation would act to blunt or negate any therapeutic effects of EGFR-TKIs that took place further upstream.
The results of this research are consistent with clinical data showing that breast cancer patients with high levels of FAM83A have a significantly lower survival rate than patients with low levels of FAM83A. However, while Bissell, Furuta and their colleagues note that a number of questions about FAM83A and other members of the FAM83 protein family remain to be addressed, the importance of these proteins as potential drug targets for therapy seems clear.
"The beauty of this study is that it not only helps explain why some breast cancer patients are resistant to EGFR-TKIs, but it also reveals a whole new family of potential oncogenes that could be a target for all types of cancer, including breast cancer," Bissell says.
Bissell says the results of this study also demonstrate the potential of using 3D phenotypic reversion assays as a new path to the discovery of more effective therapeutic drugs.
"Our 3D phenotypic reversion assay revealed the malignant phenotype, something that could not have been done with a 2D assay," she says. "This is the first time we have used our assay to discover a potential target for cancer drugs, but it shows that an assay like ours can be a powerful tool for finding new targets and therapies."