Computer model helps researchers hunt out cancer-causing mutational signatures in the genome
(Medical Xpress)—Researchers from the Wellcome Trust Sanger Institute's cancer genome project have developed a computer model to identify the fingerprints of DNA-damaging processes that drive cancer development. Armed with these signatures, scientists will be able to search for the chemicals, biological pathways and environmental agents responsible.
The computer model will help to overcome a fundamental problem in studying cancer genomes: that the DNA contains not only the mutations that have contributed to cancer development, but also an entire lifetime's worth of other mutations that have also been acquired. These mutations are layered on top of each other and trying to unpick the individual mutations, when they appeared, and the processes that caused them is a daunting task.
"The problem we have solved can be compared to the well-known cocktail party problem," explains Ludmil Alexandrov, first author of the paper from Sanger Institute. "At a party there are lots of people talking simultaneously and, if you place microphones all over the room, each one will record a mixture of all the conversations. To understand what is going on you need to be able to separate out the individual discussions. The same is true in cancer genomics. We have catalogues of mutations from cancer genomes and each catalogue contains the signatures of all the mutational processes that have acted on that patient's genome since birth. Our model allows us to identify the signatures produced by different mutation-causing processes within these catalogues."
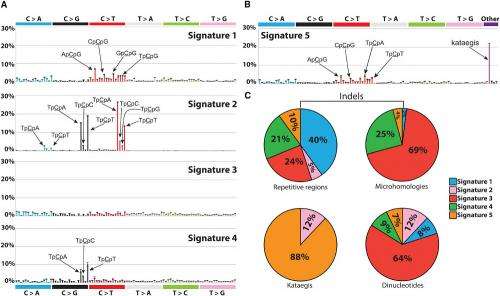
To identify individual sets of mutations produced by a particular DNA-damaging agent, the cancer genome project at the Sanger Institute simulated cancer genomes and developed a technique to search for these mutational signatures. This approach proved to be very successful. The research team then explored the genomes of 21 breast cancer patients and identified five mutational signatures of cancer-causing processes in the real world.
"For a long time we have known that mutational signatures exist in cancer," says Dr Peter Campbell, Head of the cancer genome project and co-senior author of the paper. "For example UV light and tobacco smoke both produce very specific signatures in a person's genome. Using our computational framework, we expect to uncover and identify further mutational signatures that are diagnostic for specific DNA-damaging processes, shedding greater light on how cancer develops."
The computer model offers great potential for future study: not only can the approach be applied to all forms of cancer, it can also be used to search for almost all forms of DNA damage. Depending on the type of changes the researchers want to study, the model can include single base substitutions, double nucleotide substitutions, indels, geographically localised forms of mutation such as kataegis and mutation features such as transcriptional strand bias. It is also possible that rearrangements and copy number changes (and potentially even epigenetic changes) could be incorporated into the model, providing a comprehensive overview of all the mutational processes at work.
"This new approach provides us with a valuable tool for exploring cancer genomes with a clarity and understanding that we haven't had before," says Professor Mike Stratton, Director of the Sanger Institute and co-senior author of the paper. "It will enable us to create a compendium of the mutational signatures of the many different DNA-damaging processes that operate during cancer development. This will help us understand why we get cancer by pointing to the underlying biological processes that mutate cells and cause them to become cancers, allowing us to estimate when each process took place and how much it contributed to each case."
The team is now applying the model to several hundred whole genome cancer genomes and several thousand whole-exome (protein-coding regions of the genome) cancer samples to search for more mutational signatures. These signatures will enable the team to hypothesise what the causative agents could be, enabling testing in model systems (such as yeast) to reveal the biological, chemical or environmental culprits.
More information: Alexandrov, et al., Deciphering Signatures of Mutational Processes Operative in Human Cancer. Cell Reports, 2013. DOI: 10.1016/j.celrep.2012.12.008