Researchers engineer 'protein switch' to dissect role of cancer's key players
Researchers at the University of North Carolina at Chapel Hill School of Medicine have "rationally rewired" some of the cell's smallest components to create proteins that can be switched on or off by command. These "protein switches" can be used to interrogate the inner workings of each cell, helping scientists uncover the molecular mechanisms of human health and disease.
In the first application of this approach, the UNC researchers showed how a protein called Src kinase influences the way cells extend and move, a previously unknown role that is consistent with the protein's ties to tumor progression and metastasis.
"This rationally designed control of protein conformations represents a breakthrough in computational protein design," said senior study author Nikolay Dokholyan, PhD, a professor of biochemistry and biophysics. "We now have a new tool for delineating the activities of various proteins in living cells in a way that was never before possible."
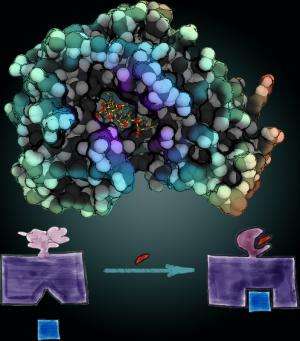
The research was published online ahead of print in the Proceedings of the National Academy of Sciences. In the study, Dokholyan created a "switch" that would make a protein wobbly and unable to do its job unless it was flipped "on" by a drug called rapamycin, which would stabilize the protein and let it perform its function.
The approach is a simpler and more reliable version of a protein engineering system pioneered three years ago by Dokholyan and Klaus Hahn, professor of pharmacology at UNC, called rapamycin regulated or RapR. In the old approach, the switching mechanism depended on two proteins and the drug. The first protein – the one the researchers wanted to study – was given the RapR modification and put in cells in tissue culture. The second protein was placed in the cells as well, but simply floated around until the addition of drug caused it to latch on to the modification in the first protein and turn it on. The problem with the approach was that some cells would have a lot of the first protein and less of the second, or vice versa.
"It became the Achilles heel of the technique, because there was variability in the results due to the different ratios between the proteins," said Hahn. "What Dokholyan was able to do, which was extremely challenging from a protein engineering standpoint, was to combine the two parts into one."
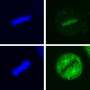
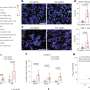
Dokholyan and his colleagues took the two proteins and broke them apart into their individual components, structures called alpha helices and beta sheets. They then rewired them together to make a whole new protein where the parts could interact with each other. When researchers compared this system, called uniRapR, with the previous approach, they found the new one gave cleaner, more reliable and more consistent results.
They then applied the technique to study Src kinase, a protein involved in the metastasis or spread of tumor cells. Scientists had postulated that Src kinase plays a role in cell motility, but previous methods have not allowed them to isolate its activity from other similar proteins.
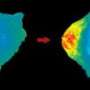
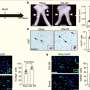
Working both in cultured human cells and in the model organism zebrafish, the researchers showed that turning on Src causes the cell to extend its edges as part of cell movement. Now that they have dissected the role of one protein, the researchers plan to look at a variety of other kinases to understand their roles in the development, progression, and spread of cancer.