How pairs of proteins collaborate to splice RNAs into cell-specific products

A team of scientists led by RIKEN researcher Yutaka Muto, in collaboration with Hidehito Kuroyanagi of Tokyo Medical and Dental University and Masatoshi Hagiwara of Kyoto University, has revealed some of the mechanisms that regulate the mysterious RNA splicing process that produces specific protein variants.
Gene expression involves transcription of DNA to messenger RNA (mRNA) followed by translation of mRNA into proteins. Although each gene is coded to produce a single corresponding protein, a mechanism known as alternative splicing makes it possible for mRNA to be translated into any number of different proteins. Each mRNA transcript consists of numerous protein-coding 'exon' segments separated by non-coding 'intron' segments. Alternative splicing is a regulated process in which splicing proteins selectively target different combinations of exons to produce a variety of possible proteins.
Many splicing proteins rely on elements known as RNA-recognition motifs (RRMs) to recognize target sequences within mRNA introns. Yet these RNA-recognition motifs are very similar to one another, and it has remained unclear exactly how splicing proteins achieve their remarkable selectivity.
Muto, a scientist from the RIKEN Center for Life Science Technologies and Musashino University who started this research while at the recently reorganized RIKEN Systems and Structural Biology Center, focused his team's attention on a pair of proteins, ASD-1 and SUP-12. These proteins are known to collaboratively coordinate alternative splicing of the mRNA encoding the signaling protein EGL-15 in the worm species Caenorhabditis elegans. EGL-15 undergoes a distinctive splicing event in muscle tissue that contributes critically to sexual development. ASD-1 is expressed throughout the worm's body, whereas SUP-12 is only produced in muscle tissue.
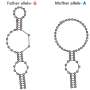
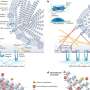
The researchers determined that SUP-12 RRM is not very selective on its own, but gets steered by ASD-1 RRM to the appropriate 12-nucleotide target sequence within the egl-15 mRNA. Detailed structural analysis using nuclear magnetic resonance (NMR) techniques revealed that ASD-1 RRM binding initiates a rearrangement in the RNA that favors SUP-12 RRM binding, resulting in a distinctive complex in which a single guanine nucleotide is essentially wedged between the two proteins (Fig. 1). Altering this single nucleotide was sufficient to eliminate the muscle-specific alternative splicing of EGL-15.
"There have been many reports that multiple splicing regulators cooperate for accurate splicing on a biochemical level, but our work is the first structural explanation for the cooperation between splicing regulatory factors at atomic resolution," says Muto. His team hopes to further determine whether such mechanisms are a general feature of cooperative alternative splicing scenarios. "We would like to advance our structural analysis to take into account the dynamic changes in the structure of the RNA molecule," says Muto.
More information: Kuwasako, K., Takahashi, M., Unzai, S., Tsuda, K., Yoshikawa, S., He, F., Kobayashi, N., Güntert, P., Shirouzu, M., Ito, T. et al. "RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing." Nature Structural & Molecular Biology 21, 778–786 (2014). DOI: 10.1038/nsmb.2870