November 18, 2014 report
Research team finds mice and humans express genes differently
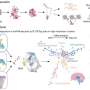
(Medical Xpress)—A large team of bio-researchers with members from the U.S. and Spain has concluded that while mice and humans have almost the same gene pool, the way those genes are expressed differ. In their paper published in Proceedings of the National Academy of Sciences, the researchers describe how they analyzed 15 different genes that were expressed in both humans and mice and found that gene expression clustered around species more so than tissue type.
Mice have of course, been used as test subjects for years—their physical makeup, it is thought, is similar enough to ours to allow for making educated guesses about how drugs and other chemicals will impact our bodies based on the way it impacts theirs. And while this model has worked well in many cases, in others it has not. In this new effort, the researchers may have found a possible clue as to why the latter occurs.
The process by which having a certain type of gene leads to the appearance of a certain trait, is known as gene expression—whereby traits can be attributable to a certain gene. Scientists have been working under the assumption that animals that are similar to us in many ways, probably experience gene expression in similar ways as well. This may not be the case however, as the researchers found that gene expression in different types of tissue in mice were more alike than same tissue types in humans. Gene expression in heart tissue in a mouse, for example, was more like gene expression in mouse liver tissue than for heart tissue in a human. In their examination, the researchers found 4,000 genes that were expressed differently by the two species, suggesting that the results found could not likely to be attributed to an artifact of the experimental process.
Previous studies on gene expression in differing species have found the opposite to be true, but the researches with this new effort suggest older techniques showed different results because of the types of tissue examined—heart, kidney, brain, liver, etc. all of which have close parallel functions in both species. The limited number of tissue types tended to skew the results.
More information: "Comparison of the transcriptional landscapes between human and mouse tissues," by Shin Lin et al. PNAS, www.pnas.org/lookup/doi/10.1073/pnas.1413624111
© 2014 Medical Xpress