September 18, 2015 report
Targeting gene interactions to kill tumor cells
(Medical Xpress)—A particular kind of genetic interaction called synthetic dosage lethality (SDL) is a promising avenue for future cancer treatment, according to a study reported in the Proceedings of the National Academy of Sciences. An international group of researchers has designed a network model of these interactions that points toward therapeutic approaches to tumors while also illuminating the value of model-based studies of metabolic processes.
An SDL is a genetic interaction in which the underexpression of gene A (A↓) combined with the overexpression of gene B (B↑) is lethal to the cell. This is particularly relevant to targeting cancer cells with over-expressed oncogenes— such oncogenes drive tumor growth, but are difficult to target directly because they are essential for cell function. Instead, the researchers propose targeting the genetic partners of oncogenes in SDL pairs.
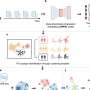
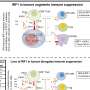
For the study, the researchers designed a biological network model as a complement to data-mining efforts used to identify SDLs. Their computational approach, called "identifying dosage lethality effects" (IDLE), predicts enzymatic SDLs from a genome-scale model of metabolism; they systematically knocked out the enzyme flux through A combined with flux increases through B and quantified the resulting cell growth reductions.
The method not only identifies SDLs that are lethal to the cell; it also pinpoints those that had significant effects on tumor growth or proliferation in clinical settings. "It is therefore of interest to focus further research toward therapeutic interventions on the basis of 'high-impact' pairs, which may have the largest beneficial effect of killing cancer cells," the authors write.
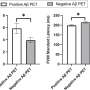
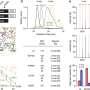
They discovered that SDLs are less frequently active in cancer cells than they expected, demonstrating that rapidly growing tumor cells select against the interactions that reduce their growth rates. Activating the high-impact SDLs was associated with smaller tumor sizes and longer patient survival. They also demonstrated that the more active SDLs were present in tumor cells, the better the patient's prognosis.
As they were conducting the first study of its kind, the researchers were particularly interested in determining which interactions were common to the greatest number of cancer types, as resulting therapeutic interventions could be applied across a larger spectrum of patients. Glycolysis is a widespread metabolic mechanism among cancer cells, and they focused on related SDLs, expecting that the down-regulation of glycolytic enzymes would have a significant result. "Intriguingly, we do find that disrupted glycogen metabolism is predicted to be the major mechanism by which hundreds of SDLs of key glycolytic enzymes exert their growth-inhibitory effect," they write.
They note that their prediction modeling is applicable beyond oncology, and could be used to identify the SDL networks of pathogenic bacteria or fungi, or applied to the engineering of SDL effects to eliminate unwanted byproducts.
More information: "Synthetic dosage lethality in the human metabolic network is highly predictive of tumor growth and cancer patient survival." PNAS 2015 ; published ahead of print September 14, 2015, DOI: 10.1073/pnas.1508573112
Abstract
Synthetic dosage lethality (SDL) denotes a genetic interaction between two genes whereby the underexpression of gene A combined with the overexpression of gene B is lethal. SDLs offer a promising way to kill cancer cells by inhibiting the activity of SDL partners of activated oncogenes in tumors, which are often difficult to target directly. As experimental genome-wide SDL screens are still scarce, here we introduce a network-level computational modeling framework that quantitatively predicts human SDLs in metabolism. For each enzyme pair (A, B) we systematically knock out the flux through A combined with a stepwise flux increase through B and search for pairs that reduce cellular growth more than when either enzyme is perturbed individually. The predictive signal of the emerging network of 12,000 SDLs is demonstrated in five different ways. (i) It can be successfully used to predict gene essentiality in shRNA cancer cell line screens. Moving to clinical tumors, we show that (ii) SDLs are significantly underrepresented in tumors. Furthermore, breast cancer tumors with SDLs active (iii) have smaller sizes and (iv) result in increased patient survival, indicating that activation of SDLs increases cancer vulnerability. Finally, (v) patient survival improves when multiple SDLs are present, pointing to a cumulative effect. This study lays the basis for quantitative identification of cancer SDLs in a model-based mechanistic manner. The approach presented can be used to identify SDLs in species and cell types in which "omics" data necessary for data-driven identification are missing.
© 2015 Medical Xpress