Screening the dark genome for disease

Researchers have developed a method to swiftly screen the non-coding DNA of the human genome for links to diseases that are driven by changes in gene regulation. The technique could revolutionize modern medicine's understanding of the genetically inherited risks of developing heart disease, diabetes, cancer, neurological disorders and others, and lead to new treatments.
The study appeared online in Nature Biotechnology on April 3, 2017.
"Identifying single mutations that cause rare, devastating diseases like muscular dystrophy has become relatively straightforward," said Charles Gersbach, the Rooney Family Associate Professor of Biomedical Engineering at Duke University. "But more common diseases that run in families often involve lots of genes as well as genetic reactions to environmental factors. It's a much more complicated story, and we've been wanting a way to better understand it. Now we've found a way."
The new technique relies on the gene-hacking system called CRISPR/Cas9. Originally discovered as a natural antiviral defense mechanism in bacteria, the system recognizes and homes in on the genetic code of previous intruders and then chops up their DNA. In the past several years, researchers have harnessed this biologic system to precisely cut and paste DNA sequences in living organisms.
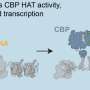
In the current study, researchers added molecular machinery that can control gene activity by manipulating the web of biomolecules that determines which genes each cell activates and to what degree.
With the new tool, Gersbach and his colleagues are exploring the 98 percent of our genetic code often referred to as the "dark matter of the genome."
"Only a small fraction of our genome encodes instructions to make proteins that guide cellular activity," said Tyler Klann, the biomedical engineering graduate student who led the work in Gersbach's lab. "But more than 90 percent of the genetic variation in the human population that is associated with common disease falls outside of those genes. We set out to develop a technology to map this part of the genome and understand what it is doing."
The answer, says Klann, lies with promoters and enhancers. Promoters sit directly next to the genes they control. Enhancers, however, which modulate promoters, can be just about anywhere due to the genome's complex 3D geometry, making it difficult to discern what they're actually doing.
"If an enhancer is dialing a promoter up or down by 10 or 20 percent, that could logically explain a small genetic contribution to cardiovascular disease, for example," said Gersbach. "With this CRISPR-based system, we can more strongly turn these enhancers on and off to see exactly what effect they're having on the cell. By developing therapies that more dramatically affect these targets in the right direction, we could have a significant effect on the corresponding disease."
That's all well and good for exploring the regions of the genome that researchers have already identified as being linked to diseases, but there are potentially millions of sites in the genome with unknown functions. To dive down the dark genome rabbit hole, Gersbach turned to colleagues Greg Crawford, associate professor of pediatrics and medical genetics, and Tim Reddy, assistant professor of bioinformatics and biostatistics. All three professors work together in the Duke Center for Genomic and Computational Biology.
Crawford developed a way of determining which sections of DNA are open for business. That is, which sections are not tightly packed away, providing access for interactions with biomachinery such as RNA and proteins. These sites, the researchers reason, are the most likely to be contributing to a cell's activity in some way. Reddy has been developing computational tools for interpreting these large genomic data sets.
Over the past decade, Crawford has scanned hundreds of types of cells and tissues affected by various diseases and drugs and come up with a list of more than 2 million potentially important sites in the dark genome—clearly far too many to investigate one at a time. In the new study, Crawford, Reddy and Gersbach demonstrate a high-throughput screening method to investigate many of these potentially important genetic sequences in short order. While these initial studies screened hundreds of these sites across millions of base pairs of the genome, the researchers are now working to scale this up 100- to 1000-fold.
"Small molecules can target proteins and RNA interference targets RNA, but we needed something to go in and modulate the non-coding part of the genome," said Crawford. "Up until now, we didn't have that."
The method starts by delivering millions of CRISPR systems loaded into viruses, each targeting a different genetic point of interest, to millions of cells in a single dish. After ensuring each cell receives only one virus, the team screens them for changes in their gene expression or cellular functions.
For example, someone researching diabetes could do this with pancreatic cells and watch for changes in insulin production. Those cells that show interesting alterations are then isolated and sequenced to determine which stretch of DNA the CRISPR affected, revealing a new genetic piece of the diabetes puzzle.
The technique is already producing results, identifying previously known genetic regulatory elements while also spotting a few new ones. The results also showed it can be used to turn genes either on or off, which is superior to other tools for studying biology which only turn genes off. Different cell types also produced different—but partially overlapping—results, highlighting the biological complexity in gene regulation and disease that can be interrogated with this technology.
"Now that we have this tool, we can go in and annotate the functions of these previously unknown but important stretches of our genome," said Gersbach. "With so many places to look, and the ability to do it quickly and robustly, we'll undoubtedly find new segments that are important for disease, which will provide new avenues for developing therapeutics."
More information: CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome, Nature Biotechnology, nature.com/articles/doi:10.1038/nbt.3853