Researchers map molecular interaction that prevents aggressive breast cancer
Researchers in Italy have discovered how specific versions of a protein called Numb protect the key tumor suppressor p53 from destruction. The study, which will be published December 21 in the Journal of Cell Biology, suggests that the loss of these particular Numb proteins makes breast cancers more aggressive and resistant to chemotherapy, but points the way toward new therapeutic approaches that could improve patient outcome by preserving p53 levels.
Cells produce several alternative isoforms of Numb by differentially processing, or splicing, the mRNA encoding Numb to include or exclude specific regions of the protein. How this alternative splicing affects Numb's various functions remains unclear.
In mammary gland stem cells, for example, Numb binds and inhibits an enzyme called Mdm2, preventing it from targeting p53 for degradation. Numb therefore stabilizes p53 and allows this tumor suppressor protein to limit stem cell proliferation. If the stem cells lose Numb, however, p53 levels plunge and the cells proliferate uncontrollably, leading to the emergence of cancer stem cells that drive the growth of breast tumors. Cancer cells that lack p53 are also more resistant to chemotherapy drugs that kill cells by damaging their DNA.
A team of researchers based in Milan set out to identify how Numb binds to Mdm2. The team was led by Pier Paolo Di Fiore of the FIRC Institute for Molecular Oncology (IFOM), the European Institute of Oncology (EIO), and The University of Milan, as well as Salvatore Pece of EIO and The University of Milan and Marina Mapelli of EIO.
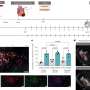
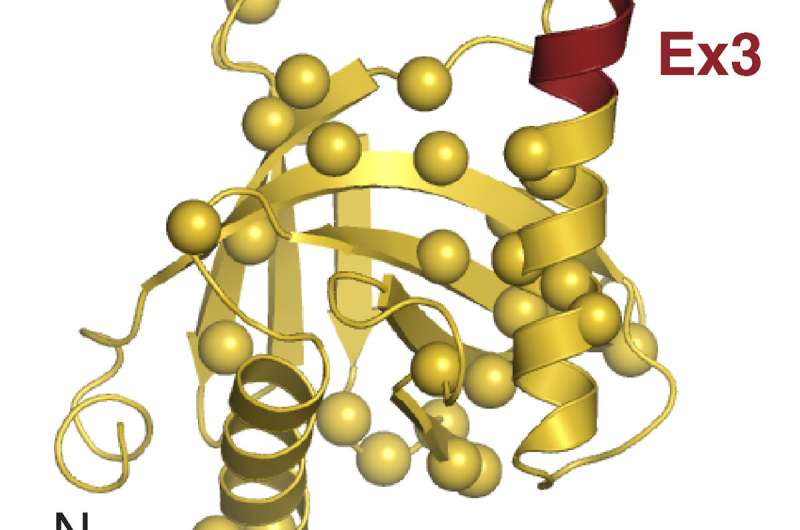
The researchers found that a small region of Numb—comprising just 11 amino acids—is responsible for binding and inhibiting Mdm2. This region is present in Numb isoforms 1 and 2 but excluded from isoforms 3 and 4. Accordingly, depleting Numb-1 and -2 from breast cancer cells reduced the levels of p53, whereas depleting Numb-3 and -4 had no effect.
The researchers then compared tumor cells isolated from multiple different breast cancer patients and found that cells expressing lower amounts of Numb-1 and -2 were more resistant to the chemotherapy agent cisplatin. Treating these cells with an Mdm2 inhibitor boosted p53 levels and increased the cells' sensitivity to cisplatin.
"We reasoned that breast cancers displaying reduced levels of Numb-1 and -2, being resistant to genotoxic agents, might also display poorer disease outcome," explains Pece.
The team therefore analyzed the case history of 890 breast cancer patients and found that low Numb-1 and -2 levels correlated with an increased risk of aggressive, metastatic disease, particularly for the luminal subtype of breast cancers, which tend to retain a normal, functional copy of the p53 gene.
"Our results show how Numb splicing specifically impacts the regulation of p53 and breast cancer prognosis," Mapelli says.
"We hope that it will be possible to exploit the knowledge of the molecular basis of the Numb-Mdm2 interaction in the rational design of molecules that can mimic the crucial region in Numb and inhibit Mdm2 to relieve p53 dysfunction in Numb-defective breast cancers," Di Fiore says.
More information: Colaluca et al., Journal of Cell Biology (2018). DOI: 10.1083/jcb.201709092