Protein research: Supposed disorder is not disorder after all

Many, but not all, proteins in a living cell have a defined three-dimensional structure. The interrelationship between the structure and function of proteins is the focus of many research initiatives that extend to the development of innovative drugs.
"However, based on recent research results, it is predicted that at least 30 percent of all proteins in cells containing a nucleus are partially or even completely unstructured," says Raphael Stoll, head of the Biomolecular Spectroscopy research group. Because of this remarkable feature, these proteins have special, sometimes crucial, functions in both healthy and disease-causing processes. These include, for example, the regulation of the cell cycle, the transmission of biological signals, and the development of cancer or neurodegenerative diseases such as Alzheimer's or Parkinson's disease.
One of these seemingly disordered proteins is the high-mobility group protein A1a (HMGA1a). It is highly abundant in the cell nucleus and is important for embryonic development, cell differentiation, and is also involved in the development of the uncontrolled cell proliferation of neoplasia.
The first full-length structural model
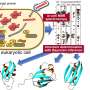
The Bochum-based research team shows for the first time that the HMGA1a protein does not adopt completely random forms, but dynamic and more compact structures. This enabled the researchers to create the first full-length structural model of the HMGA1a protein.
They were also able to describe the structural effects of the phosphorylation of the HMGA1a protein on its function. The attachment of phosphoryl groups alters the function of many proteins and they are thus switched on or off. Phosphoryl groups can also influence the protein's ability to bind to other cell components. HMGA1a binds to DNA. This process is very important for its biological mode of action as HMGA1a, for example, is involved in regulating the formation of RNA and reorganising chromosomes.
Structure and binding probabilities change
The researchers applied nuclear magnetic resonance spectroscopy, which is able to provide information not only on the structure but also on the dynamics of proteins. "Our results show that the dynamic and compact structures of this protein depend on its phosphorylation state," reports Raphael Stoll. Within the cell, the HMGA1a protein is phosphorylated by casein kinase 2. This has an effect on the electrostatic network in the HMGA1a protein and thus changes the dynamic structural ensemble of this protein. Further experiments revealed that these changes even affect the ability of the HMGA1a protein to bind to its natural target sequence in the DNA of the cell nucleus.
More information: Phosphorylation orchestrates the structural ensemble of the intrinsically disordered protein HMGA1 and modulates its DNA binding to the NFKB promoter, Nucleic Acids Research, 2019, DOI: 10.1093/nar/gkz614/5538012