A computational model to understand the dynamics of tuberculosis lesions within the lungs
The researchers of the Experimental Tuberculosis Unit of the Germans Trias i Pujol Research Institute (IGTP), led by Pere-Joan Cardona, have published the first results of a computational model that aims to reproduce the dynamics of tuberculosis lesions in a virtual lung. The model has been developed within the 3Rs Program of the Centre for Comparative Medicine and Bioimaging (CMCiB) of the IGTP, supported by the "la Caixa" Foundation.
Martí Català and Clara Prats, researchers from the Department of Physics of the Universitat Politècnica de Catalunya, Barcelona Tech (UPC) at the CMCiB, and Daniel López-Codina, Sergio Alonso and Joaquim Valls, who are also UPC researchers, took part in the study. Català, the first author of the paper, led the in silico trial of the program. Its results have been published in the journal PLOS Computational Biology.
Today, tuberculosis (TB) is still among the world's top 10 causes of death, killing 1.6 million people in 2017. Tuberculosis is caused by Mycobacterium tuberculosis, a bacterium that infects pulmonary alveoli. However, 90% of people infected never develop the active disease. Not knowing the main factors that trigger the disease in the remaining 10% of cases is one of the main obstacles to its eradication. This study aims to understand the mechanisms that allow the disease to remain latent, especially those related to endogenous reinfection.
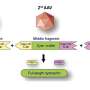
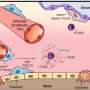
The researchers have started from the hypothesis that endogenous reinfection plays an important role in maintaining latent infection. "So we have developed an agent-based model that describes the growth, fusion and proliferation of tuberculosis lesions in a computational bronchial tree, built from an iterative algorithm that generates bronchial tubes and bifurcations within a 3-D volume of the lung surface area," explains Clara Prats, a researcher at the UPC's Department of Physics and a member of the Computational Biology and Complex Systems Group.
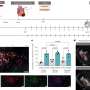
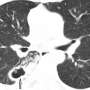
The model is based on experimental data from computed tomography scans of five mini-pigs in the initial stages of infection. The lung surface area of the mini-pigs has been obtained from these images in order to generate the computational lung. The size and location of each of the TB lesions has also been obtained, which allowed the researchers to set the parameters of the agent-based model.
"The result is a model that allows us to reproduce and understand the experimental data on the computer. We have been able to show an important relationship between the final number of tuberculosis lesions and the frequency of endogenous reinfection and lesion growth," says Martí Català.
The model has also been used as an in silico experimental platform to explore the transition from latent infection to active disease, identifying the main triggers: elevated inflammatory response and the combination of a moderate inflammatory response with low respiratory amplitude. "This study is important because the lung structure of mini-pigs is very similar to that of humans, so this model will allow us to make predictions for future actions such as new biomarkers, preventive strategies and therapies for tuberculosis in humans," explains Pere-Joan Cardona, the co-author of the project.
In silico or computational models
The use of animal models, known as comparative medicine, has been one of the main keys to the huge developments in medicine in the 20th century and particularly in research in the fields of health and life sciences. It remains a necessary and mandatory step for research in these fields, especially for developing new treatments and drugs. New technologies have allowed for the refinement of this practice, which is the main purpose of the research facilities and projects at the Centre for Comparative Medicine and Bioimaging (CMCiB).
The Centre promotes research based on the 3Rs: replace, reduce and refine. The use of computational tools for simulation, as in this case, is a clear example. "Less is more in comparative medicine. Our group has been using alternative models for many years in preclinical research, such as the Drosophila model for tuberculosis research, a paradigmatic example. Now, the possibilities offered by computer-generated models are enormous," explains Cardona, the principal investigator of the Experimental Tuberculosis Unit and the scientific director of the CMCiB. "In this case, we have obtained reliable results by generating a model using a small number of animals and bioimaging tools. Now we will not need more animals; the model is in the computer and we will be able to use it for many future studies," he says.
More information: Martí Català et al. Modeling the dynamics of tuberculosis lesions in a virtual lung: Role of the bronchial tree in endogenous reinfection, PLOS Computational Biology (2020). DOI: 10.1371/journal.pcbi.1007772