Individual donor strains do not show link between fecal microbe transplantation and successful melanoma immunotherapy
The mix of microbes in the gut can shape a person's immune response. In immunotherapy for melanoma, for example, the composition of the gut microbiota correlates with efficacy of anti-PD-1 therapy in animal models and cancer patients.
One clinical trial published this year
in Science put
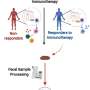
Researchers at the University of Alabama at Birmingham Marnix E. Heersink School of Medicine now have used the fecal metagenomic DNA sequencing data set from that study—as well as the DNA sequencing data sets from two other studies—to see whether specific donor strains established themselves in the recipient patients and whether those donor strains correlated with successful treatment outcomes.
The researchers, Hyunmin Koo, Ph.D., and Casey Morrow, Ph.D., are able to identify specific single strains of a bacterial species using a microbiome "fingerprint" method developed at UAB. The method uses powerful genomic tools and supercomputers to analyze massive amounts of genetic data from metagenomic sequencing of fecal samples.
This novel bioinformatics technique has previously shown, among other studies, the emergence of new microbial strains after obesity surgeries and suppressive antibiotics, as well as the stable persistence of donor microbial strains for as long as two years after fecal microbe transplantation to treat recurrent Clostridium difficile infections.
Now, in a study published in BMC Microbiology, Koo and Morrow report that the individual variation among the melanoma patients, with respect to fecal-dominant donor microbes in recipient patients after fecal microbe transplantation, did not correlate with responses to anti-PD-1 therapy.
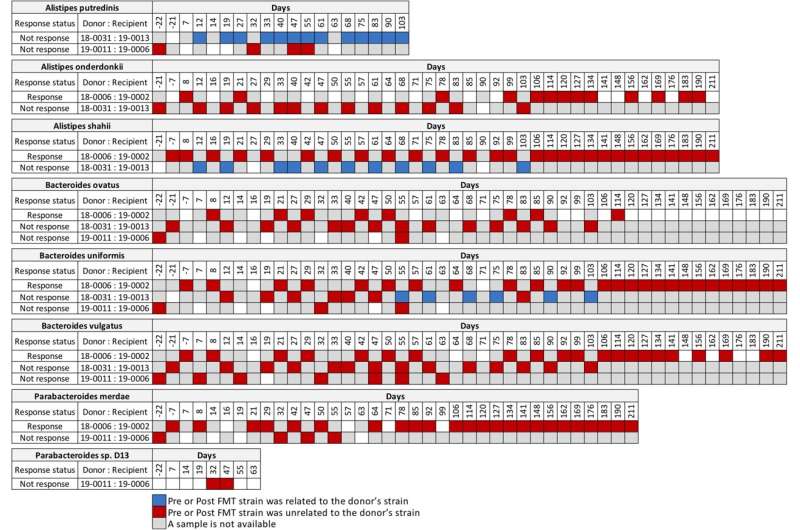
In detail, they analyzed a metagenomic sequencing data set published by Baruch et al. in Science in 2021 and found that the presence of commensal donor microbes in feces of recipient patients after fecal microbiota transplantation did not correlate with the patient response to immunotherapy.
They also analyzed the metagenomic sequencing published by Davar et al. in Science in 2021. "From the Davar et al. data set, we found four patients that responded to immunotherapy had donor related strains post-fecal microbe transplantation, while two patients that did not respond to the immunotherapy also had donor strains post-fecal microbe transplantation," Morrow said. "Importantly, we identified no donor microbes in the feces of one recipient, post-fecal microbe transplantation, who responded to immunotherapy."
Looking at longitudinal sequencing data sets from Davar et al. that analyzed fecal samples up to 18 months after transplantation, two patients who responded to immunotherapy revealed both donor and recipient strains at different times post-fecal microbe transplantation. This ongoing competition for fecal dominance over time between donor and recipient strains is further support that the link between donor microbes in the feces of recipients and response to immunotherapy is incongruent, Morrow says.
Koo and Morrow also analyzed the metagenomics data set from a study by James et al. in Nature Immunology in 2020. This research analyzed the microbiota that are associated with the mucosa of the intestinal epithelium of the normal human cecum, transverse colon and sigmoid colon to gain insights into the microbe interaction with the human immune system.
The UAB analysis showed considerable strain sharing between the cecum-transverse colon and the cecum-sigmoid colon for two bacterial species. The cecum is at the beginning of the colon, where it connects with the small intestine. The transverse colon is at the middle of the colon, and the sigmoid colon is at the end, near the rectum.
"Thus, these results demonstrate strain sharing occurs for commensal microbial strains in the intestinal epithelium throughout the entire length of the normal colon," Morrow said. "This suggests a large community structure that extends throughout the entire colon that could interact with the immune system and impact the capacity of donor microbes derived from fecal microbe transplantation to facilitate the response to immunotherapy." These niches may help the recipient resist the establishment of donor microbial strains.
"The results of our strain-tracking analysis provide evidence that the conditions used for transplant of donor microbes into existing microbial communities of the recipient do not ensure comprehensive replacement of the recipient microbes with donor microbes, and this lends new insights to explain the inter-individual variability of fecal microbe transplantation to enhance immunotherapy," Morrow said.
More information: Hyunmin Koo et al, Incongruence between dominant commensal donor microbes in recipient feces post fecal transplant and response to anti-PD-1 immunotherapy, BMC Microbiology (2021). DOI: 10.1186/s12866-021-02312-0