New pediatric tumor identifications could help predict chemo response
A new UCSF study sheds light on the diversity within the most common type of pediatric liver tumor and suggests a way forward for more precise chemotherapy treatment.
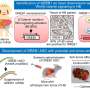
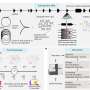
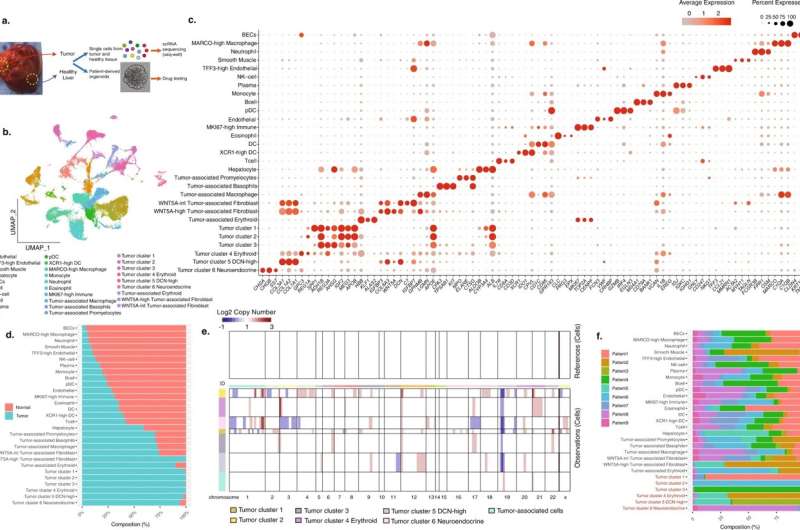
The study, published in Nature Communications, used single-cell transcriptomic techniques to analyze hepatoblastoma specimens from infants and children under age four. From nine samples, the researchers identified five hepatoblastoma tumor signatures that may account for the heterogeneity in this cancer, and that may predict responses to chemotherapy treatments.
"Many pediatric tumors originate from an embryonic cell and some are bland in terms of their mutational burden," said study co-author Amar Nijagal, assistant professor in the Division of Pediatric Surgery at UCSF Benioff Children's Hospital, San Francisco. "The fact that this tumor has significant heterogeneity made us question what was driving this heterogeneity, and we wanted to learn more."
Researchers from three different labs at UCSF worked on the study, combining expertise in pediatric surgery, hepatology, and genomics, which facilitated the rare opportunity to analyze tumors directly from the operating room, Nijagal said.
"With this combined expertise in one place at UCSF, we can foresee a situation in which a child presents with a liver tumor; we biopsy it; we understand its tumor signatures; we grow and test therapeutics in a petri dish; and we use what we find to treat the patient," Nijagal said.
Aside from offering hope for better treatment, studying hepatoblastoma provides a window into how tissues develop normally, Nijagal said.
"Normal development and cancer are two sides of the same coin," Nijagal said. "Here you have a tumor that is not acquired, as many adult tumors are from something like smoking or excess UV exposure. Instead, something goes wrong during development to set off these hepatoblasts towards developing into a malignancy. So we are interested in learning what are the signals—such as those from immune cells—that maintain normal development."
More information: Hanbing Song et al, Single-cell analysis of hepatoblastoma identifies tumor signatures that predict chemotherapy susceptibility using patient-specific tumor spheroids, Nature Communications (2022). DOI: 10.1038/s41467-022-32473-z