This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Identifying where HIV sleeps in the brain
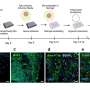
![Microglia HIV-1 ISs map to introns of highly transcribed genes recurrently targeted in T cells(A) Normalized chromosomal distribution of the integration events for CD4+ T cells, MDMs, and microglia. Integration density is calculated for each cell type as the number of integrations per chromosome per total number of integrations divided by chromosome length.(B) Bar plot showing the percentage of ISs falling into promoters (microglia, 20.1%; MDMs, 25.7%; CD4+ T cells, 20.2%; from the transcription start site [TSS] to 2 kb upstream), 3′ UTRs (microglia, 2%; MDMs, 2.4%; CD4+ T cells, 2.3%), 5′ UTRs (microglia, 0.1%; MDMs, 0.1%; CD4+ T cells, 0.14%), exons (microglia, 1.9%; MDMs, 2.5%; CD4+ T cells, 1.6%), introns (microglia, 60.2%; MDMs, 56.3%; CD4+ T cells, 57.2%), downstream (microglia, 0.9%; MDMs, 1.1%; CD4+ T cells, 0.9%; from the transcription end site [TES] to 3 kb downstream), and distal intergenic regions (microglia, 14.5%; MDMs, 11.7%; CD4+ T cells, 17.3%).(C) Shared integrations in CD4+ T cells, MDMs, and microglia. Shown is a comparison of the genes between CD4+ T cells (n = 5,199), MDMs (n = 750), and microglia (n = 2,719) with at least one integration in their gene bodies.(D) Bar plot representing the percentage of genic ISs according to their expression levels determined by RNA-seq into high, mid, low, or no expressed genes in microglia and CD4+ T cells compared with the global gene expression levels in each cell type. Credit: Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112110 Where HIV sleeps in the brain](https://scx1.b-cdn.net/csz/news/800a/2023/where-hiv-sleeps-in-th-1.jpg)
The human immunodeficiency virus HIV-1 is able to infect various tissues in humans. Once inside the cells, the virus integrates its genome into the cellular genome and establishes persistent infections. The role of the structure and organization of the host genome in HIV-1 infection is not well understood.
Using a cell culture model based on brain immune microglia cells, an international research team led by scientists from Heidelberg University Hospital and the German Center for Infection Research (DZIF) now defined the insertion patterns of HIV-1 in the genome of microglia cells.
An infection with the human immunodeficiency virus (HIV) is in 99,9% of cases still an incurable disease. This is because the virus remains dormant for a long time in the genome of infected cells, making it invisible and inaccessible to the immune system and antiviral drugs. The pathways that HIV-1 takes to remain hidden in the host cell genome have been studied primarily in blood CD4+ T cells—the main target cells of the virus.
However, HIV-1 is capable of infecting other immune cells in different organs, where it establishes stable reservoirs. One such viral sanctuary is the brain, where the virus infects mostly microglia immune cells, often causing neuroinflammation and symptoms of HIV-1 associated neurocognitive disorder (HAND).
Questions related to HIV-1 infection and replication in the brain have been very difficult to resolve because studies in patients are limited in their ability to monitor the virus in the brain. An international team led by Dr. Marina Lusic from Heidelberg University Hospital and the DZIF now succeeded in developing HIV-1-infection models in human microglia cell cultures.
Establishment of the models allowed for the first time to investigate the insertion of the HIV-1 genome in that of microglia cells. Genomic insertion results in the silencing of the viral genome, leading to the so-called "sleeping virus" phenotype.
The researchers, who recently published their findings in Cell Reports, used the microglia cell models to determine HIV-1 integration sites and associate them with structural and regulatory elements of the chromatin.
"Modeling HIV-1-infections of the brain immune cells using the microglia cell cultures, we discovered a stronger correlation between a cellular chromatin factor and the sleeping virus phenotype," says the paper's first author Mona Rheinberger.
"This protein, called CTCF, is one of the most important architectural proteins of the cellular genome, involved in chromatin folding and packaging inside the cells. Our findings indicate that CTCF is shaping the HIV-1 genomic insertion profiles in microglia, thus contributing to the latent infection state."
"These studies in cell culture models are extremely important to understand how the virus can be targeted in different parts of the body, where it can remain hidden or cause neurological disorders even under current therapeutic regimens," Dr. Lusic concludes.
More information: Mona Rheinberger et al, Genomic profiling of HIV-1 integration in microglia cells links viral integration to the topologically associated domains, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112110