March 7, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
The impact of cohesin mutations: Insights into a hidden regulator of alternative splicing in leukemia
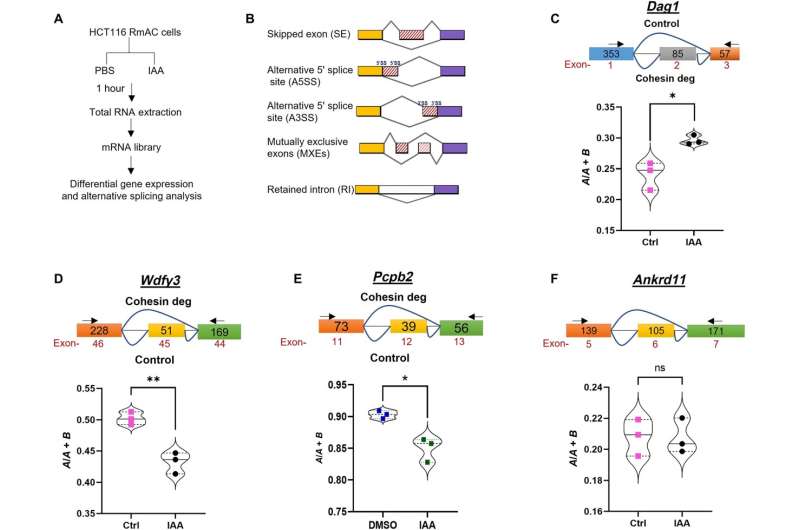
A new role for cohesin in regulating alternative splicing has been identified, providing insights into splicing mutations in human disease.
Cohesin is known to play a key part in transcription, and now researchers believe it does more than that. A new study published in the journal Science Advances found that causal relationships with known splicing factors, correlations in public data sets of unique splicing patterns and degradation of cohesin resulted in marked changes in splicing independent of effects on transcription.
Alternative splicing is a routine activity that takes place during the production of proteins in humans. The genome is packed full of basic functional codes for making proteins, but the body relies on more than what is coded to function. There are mechanisms that augment RNA to form multiple proteins that carry out different tissue-specific roles. Sort of like how a basic muffin recipe can be turned into a variety of tasty treats, alternative splicing adds and rearranges the RNA ingredients to form new proteins.
There are known factors involved in alternative splicing, and cohesin was not on that list. However, a disease where cohesin mutations have been observed at frequencies as high as 20% is acute myeloid leukemia (AML). AML is frequently associated with significant changes in patterns of alternative splicing. This association was intriguing enough for a closer look.
National Cancer Institute researchers analyzed two independent publicly available sets of RNA-sequenced data from AML patient samples. They looked at samples with mutations in known splicing factors but no cohesin mutations. This would be the previously expected cause of incorrect alternative splicing. Researchers compared them to splicing patterns in samples of AML patients without mutations in either cohesin or splicing factors (a sort of wild-type control group). And a third grouping of splicing patterns in samples with cohesin mutations but without mutations in splicing factors. If cohesin wasn't a factor, this should be no different from the wild type.
The smoking gun
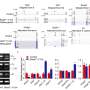
According to the study, analyses revealed unique patterns of splicing associated with cohesin mutations that were not observed in the other datasets and significant differences in splicing patterns when the AML samples with cohesin mutations (but no splicing factor mutations) were compared to either those with splicing factor mutations.
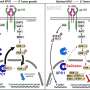
In a series of experiments where a known splicing factor, BRD4, was selectively included or excluded from cohesin-containing cells, researchers observed distinct patterns of alternative splicing regulated by cohesin or BRD4 alone and in combination. These results define a previously unknown role for cohesin in regulating alternative splicing, both alone and with BRD4 through direct contact with splicing machinery components.
To further document that cohesin mutations directly affect alternative splicing, researchers analyzed two clones of mouse embryonic stem cells engineered with a single-point mutation (one often observed in AML patients). Researchers observed reduced interactions with splicing factors and altered splicing patterns, demonstrating causality.
The findings of the research team identify a role for cohesin in regulating alternative splicing in both normal and leukemic cells and provide insights into the role of cohesin mutations in human disease. As with any good study, more research is now needed to investigate the influence of cohesin mutations on other cancers and illnesses where its role has previously gone unnoticed.
More information: Amit K. Singh et al, Cohesin regulates alternative splicing, Science Advances (2023). DOI: 10.1126/sciadv.ade3876
© 2023 Science X Network