This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers use 'deep sequencing' to identify previously undescribed genetic variants in vascular anomalies
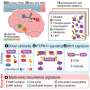
Researchers from Children's Hospital of Philadelphia (CHOP) recently discovered that extremely thorough "deep sequencing" of the genome in tissue samples and cell-free DNA of patients with potentially life-threatening vascular anomalies captured several genetic variants related to disease that were not captured with conventional genetic sequencing methods. More than 60% of patients saw an improvement in their condition after being placed on targeted therapies related to these newly found genetic variants. The findings were published June 1, 2023, in the journal Nature Medicine.
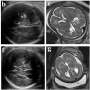
Vascular anomalies describe a variety of conditions that affect the veins, arteries and lymphatic system that can be classified as either vascular tumors (benign or malignant) or vascular malformations. While certain vascular anomalies naturally resolve over time, others can cause visible deformities, impede critical organ functions such as swallowing or breathing, or cause severe pain. Some vascular anomalies can even be life-threatening.
In a prior study also published in Nature Medicine, CHOP researchers were the first to discover a genetic variant that was responsible for a vascular anomaly affecting the lymphatic system, which allowed the clinical team to repurpose an existing drug to treat a patient that improved his breathing capacity and dramatically reduced swelling of his legs, side effects of his condition.
The research team suspected that other patients affected by vascular anomalies might also have mutations driving diseases that would benefit from targeted therapies. However, a lack of access to affected tissue samples or insufficient genomic sequencing information meant that the gene variants responsible for these issues may not be captured by conventional genetic testing.
"While some patients have inherited variants that you can find in a blood sample, about 90% of patients with vascular anomalies have acquired somatic mutations, or mutations that are not inherited, which are usually present in very low frequencies and only in certain cell or tissue types," said senior study author Hakon Hakonarson, MD, Ph.D., director of the Center for Applied Genomics and co-principal investigator of the Comprehensive Vascular Anomalies Frontier Program at CHOP. "In many cases, the disease-causing variant in the mutated gene of interest is present in frequencies of less than 1%, which makes them hard to detect with conventional sequencing approaches."
To better capture the underlying genetics behind these more severe vascular anomalies, researchers studied DNA from CD31+ cells or cell-free DNA isolated from lymphatic fluid or plasma from a cohort of 356 patients, including 104 with primary complex lymphatic anomalies.
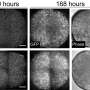
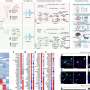
The isolated DNA underwent deep sequencing, which involves repeatedly sequencing certain areas of interest in the genome several times, and uncovered several somatic variants that were identified for the first time. This deep sequencing achieved a variant allele frequency of 0.15%, meaning that deep sequencing could detect variants that had a frequency as low as 0.15% in a particular specimen.
By identifying these variants, the researchers and clinical team were able to provide a molecular diagnosis, including previously undescribed genetic causes, in 41% of patients with primary complex lymphatic anomalies and 72% of patients with vascular malformations. As a result, 69 patients received or planned to receive a new medical therapy, and 63% of patients experienced marked improvement in their symptoms.
"The ability to link a patient's phenotype to the causative genotype of the vascular anomaly has been critical for patients," said study author Denise Adams, MD, a pediatric hematologist-oncologist and Director of the Comprehensive Vascular Anomalies Program at CHOP. "This has enabled treatment with directed medical therapy that has significantly improved the quality of life of our patients. We are fortunate to work with a wonderful interdisciplinary team that has helped to move this forward for our patients."
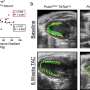
"Importantly, our study comprehensively demonstrated the bedside to bench and back approach—from the molecular studies that found the low allele frequency variants to the functional studies in organoids and zebrafish that ultimately benefited the patients by directing medical therapy," said study co-leader Sarah Sheppard, MD, Ph.D., a tenure track investigator at the Eunice Kennedy Shriver National Institute of Child Health and Human Development and clinical geneticist for the Comprehensive Vascular Anomalies Program at CHOP.
"Our findings pave the way for future applications of cfDNA technology to be an innovative, non-invasive molecular diagnostic for all patients with vascular anomalies," said study co-leader Dong Li, Ph.D., an assistant professor within the Center for Applied Genomics at CHOP. "We believe the time is right to transform the understanding of these complex diseases and identify and test new therapies for these life-threatening and life-altering conditions."
More information: Li, Sheppard et al, Genomic profiling informs diagnoses and treatment in vascular anomalies, Nature Medicine (2023). DOI: 10.1038/s41591-023-02364-x